

Aust J Crop Sci. 18(10):666-672 (2024) | ISSN:1835-2707
https://doi.org/10.21475/ajcs.24.18.10.p73
Danielle Rezende Vilela1, Édila Vilela de Resende Von Pinho*1, Wilson Vicente Souza Pereira1, Edlânia Maria de Souza1, Renzo Garcia Von Pinho2, Heloisa Oliveira dos Santos1, Viviana Ramírez Rios1
1Setor de Sementes/Departamento de Agricultura. Universidade Federal de Lavras. Rotary Roundabout Professor Edmir Sá Santos, s/n • P.O. Box 3037 • ZIP Code 37203-202 • Lavras/MG, Brazil
2Setor de Grandes Culturas/Departamento de Agricultura. Universidade Federal de Lavras. Rotary Roundabout Professor Edmir Sá Santos, s/n • P.O. Box 3037 • ZIP Code 37203-202 • Lavras/MG, Brazil. Email
*Corresponding author: Édila Vilela de Resende Von Pinho 
ORCID: 0000-0001-5276-2806.
Abstract: The efficacy of a breeding program hinges on its ability to swiftly select desirable lines, as the expression of some key genes is a rapid and efficient marker. Although a wide range of genes are involved in water deficit tolerance, their expression is of considerable interest in maize breeding programs. Thus, this research aimed to analyse the expression of several water deficit-related genes in two contrasting maize lines. Two contrasting lines for drought tolerance (L91-T – tolerant and L24-NT – nontolerant) were sown and grown at two densities—60 and 120 thousand plants/hectare. Leaves, stems, and adventitious roots were collected from these plants for the assays. The relative expression of ZmDREB2A/2.1S (LOC732788), ZmPP2C (LOC542176), CAT3 (LOC542370), and SOD (LOC100136885) was analysed through qRT‒PCR. Independent of the density, higher expression of ZmDREB2A/2.1S was observed for the tolerant line for all tissues collected. For ZmPP2C, greater expression was detected in the nontolerant line. The expression of both genes was greater in the leaves of 120 thousand plants/hectare. The expression of antioxidant system genes (CAT and SOD) was greater for the nontolerant line than for the nontolerant line at 120 thousand plants/hectare. In the search for potential markers for maize water deficit tolerance, with correct adaptations and further studies, the expression of ZmDREB2A/2.1S and ZmPP2C may be used for tolerant and nontolerant lines, respectively.
Keywords: Zea mays. Drought. Plant breeding. transcription factors. Water deficit.
Although water deficit tolerance is a highly important trait in plant breeding programs, the underlying mechanism is complex, involving a wide number of genes, and has yet to be fully understood (Verma, 2016). To identify new and rapid methods for line selection in breeding programs, studies have highlighted the potential use of gene expression as a molecular marker for these aims (Roy et al., 2011). In this sense, understanding the molecular basis of tolerance to water deficit and identifying response mechanisms and genes can be useful for helping breeders develop plants with greater tolerance to this abiotic stress.
Many studies have investigated changes in plant gene expression in response to stress, with changes related to the severity and type of stress, tissue, phenological stage, and genotype (Fritsche-Neto et al., 2011; Pérez-Alfocea et al., 2011). Two gene groups can be involved in the plant stress response: the first group included genes encoding proteins related to signal transduction, such as the bZIP family gene ZmBZIP72 and the AP2/ERF family members ZmDREB1 and ZmDREB 2 (Leng and Zhao, 2020), and the second group included genes related to antioxidant system enzymes, such as superoxide dismutase, catalase, and glutathione peroxidase (Wei et al., 2022).
Signal transduction genes, such as DREB, are proteins that are reportedly linked to drought-responsive elements (transcription factors) and are widely used in transgenic plants for water deficit tolerance (Zhang et al. 2006). These genes are expressed mainly at the beginning of the gene regulation process, initiating a process that ends with stress-related gene expression (Nepomuceno et al., 2000; Benko-Iseppon et al., 2012). The DREB gene has been largely studied in Arabidopsis thaliana, as well as in other species, in response to water deficit, such as rice (Chen et al., 2008) and wheat (Maruyama et al., 2009), which are reportedly linked to water stress. Similarly, protein phosphatase 2C (PP2C) has been linked to signal transduction in response to water deficit (Marques et al., 2020; Santos et al., 2021)
In response to water deficit, metabolism may result in the production and accumulation of reactive oxygen species (ROS), which can originate from the intensification of respiration (Choudhury et al., 2017). These molecules are extremely reactive and may result in the oxidation of any molecule around the ROS and damage to cellular membranes, proteins, RNA, and DNA. To avoid this damage, cells have developed an antioxidant system that includes a set of enzymes responsible for eliminating ROS, such as
| $NRE = \ \frac{{RE}_{Sample}}{{RE}_{Control}}$ | (1) |
|---|---|
| In which $RE = \ \frac{{Ex}_{Interest}}{{Ex}_{Reference}}$ | (2) |
| In which Ex =Ef−ΔCt | (3) |
| And ΔCt = CtWitness− CtSample | (4) |
| Equation 1. Formula used to calculate the gene expression in maize. Captions: NRE: Normalized Relative Expression; RE: Relative Expression; ExInterest: expression of the target gene (gene of interest). ExReference : expression of the reference gene; Ef: primer efficiency; Ct: cycle threshold. The expression of the reference gene consisted in the average of both reference genes used on this research. | |
superoxide dismutase, catalase, and peroxidases (Das and Roychoudhury, 2014).
In corn plants exposed to water deficit, overexpression of the ZmPP2C gene increases plant sensitivity and, consequently, reduces tolerance to water stress (Liu et al., 2013). Similarly, after evaluating the functions of the phosphatase PP2C in maize seedlings, He et al. (2019) reported that ZmPP2CA2 and ZmPP2C_A6 play negative roles in stress responses to water deficit. In summary, the main strategy for maize breeding needs to be associated with conventional breeding and molecular techniques for genetic engineering. These studies are essential for increasing breeding program efficiency.
Additionally, as noted by Marchin et al. (2020), simulating water deficit in the field poses numerous challenges, making the process complex. Currently, researchers are actively seeking more practical methods for simulating drought. However, studies on plant density have indicated that while increasing the number of plants per hectare can increase yields up to a certain threshold, higher densities may increase evapotranspiration, resulting in reduced water availability (Magaia et al., 2017; Hérnandez et al., 2020). Consequently, there is a risk of water stress in plants (Echarte et al., 2019), as observed by Zhang et al. (2019) in maize, where densities above 60 thousand plants per hectare lead to drought stress. Taking this into account, some studies in the field have explored the application of higher plant densities beyond the optimal density to simulate drought stress in plants (Abreu et al., 2017; Sangoi et al., 1996; Monneveux and Ribaut, 2006).
Therefore, studies involving the identification of candidate genes and the expression of transcripts related to water deficit tolerance are important for understanding the mechanisms of this tolerance and for the selection of tolerant genotypes. Thus, the objective of this work was to evaluate the expression of candidate genes in the adventitious roots, stems, and leaves of two contrasting corn lines for water deficit tolerance and for development under different population densities.
Results
DREB2A/2.1S
For DREB2A/2.1S, as we compared the lines, the expression was greater in 91 (tolerant) in all tissues than in line 24 (nontolerant); this result was observed for both densities (Figure 1). For line 91, independent of density, higher expression was detected on the leaves, intermediate expression was detected on the stalks, and lower expression was detected on the roots. However, for line 24, the expression was greatest in leaves, followed by stalks and roots, at a density of 60 thousand plants.ha-1, and the pattern changed to a density of 120 thousand plants.ha-1, with stalks having the highest expression, followed by leaves and roots having the lowest expression (Fig. 1).
ZmPP2C
The ZmPP2C expression pattern was the opposite of that observed for DREB2A/2.1S (Figs. 1 and 2). Higher expression was detected for line 24 (nontolerant), whose leaf tissue had a greater average density of 60 thousand plants.ha-1, and stalks had a value of 120 thousand plants.ha-1 (Fig. 2). By comparing tissues, leaves had higher expression of ZMPP2C and lower expression on stalks for both lines at 60 thousand plants.ha-1, while at 120 thousand plants.ha-1, higher expression was detected in leaves for line 24, and in roots, the expression was lower. For line 91, at this density, the leaves and roots had greater expression, and the stalks had lower expression (Fig. 2).
Superoxide Dismutase
In contrast to the previous genes, SOD was not expressed in any clear pattern (Fig. 3). Except for roots at a density of 60 thousand plants.ha-1, higher expression was found for line 24 than for line 91. The expression patterns of the two lines differed depending on the density. At 60 thousand plants.ha-1, higher expression was detected in the roots of line 91 and in the leaves of line 24. At a density of 120, higher expression was detected in both the leaves and roots of the plants (Fig. 3).
Catalase
No clear pattern of CAT3 expression was found in our experiments (Fig. 4). For both the lines and densities, the leaves had greater CAT3 expression. For line 91, under both densities, the expression was lower for stalks, while for line 91, lower values were found for roots at a density of 60 thousand plants.ha-1 and stalks at 120. Higher CAT3 expression was detected in line 91 at 60 thousand plants ha-1 for all tissues except stalks. At 120 thousand plants.ha-1, higher expression was detected for line 91 on roots and stalks, while for leaves, line 24 had higher values.
Discussion
The effects of increasing maize density have been studied and reported with the aim of improving crop production (Hernández et al. 2020). Such effects have been shown in studies by Winans et al. (2021), Magaia et al. (2017), and Sani et al. (2008), whose results indicate that the highest densities for the regions where the research was conducted. Maize yields are reported to increase, together with water usage efficiency, when density increases (Al-Kaisi and Yin, 2003); however, this effect can be observed only for environments in which water availability is not limited (Magaia et al., 2017; Hernández et al., 2020). In these cases, increasing density will result in greater evapotranspiration and consequently a decrease in soil water availability and ultimately drought stress for plants (Echarte et al., 2019). Therefore, increasing densities can be used as an alternative to simulate drought in maize (Marchin et al., 2020; Abreu et al., 2017; Monneveux and Ribaut, 2006).
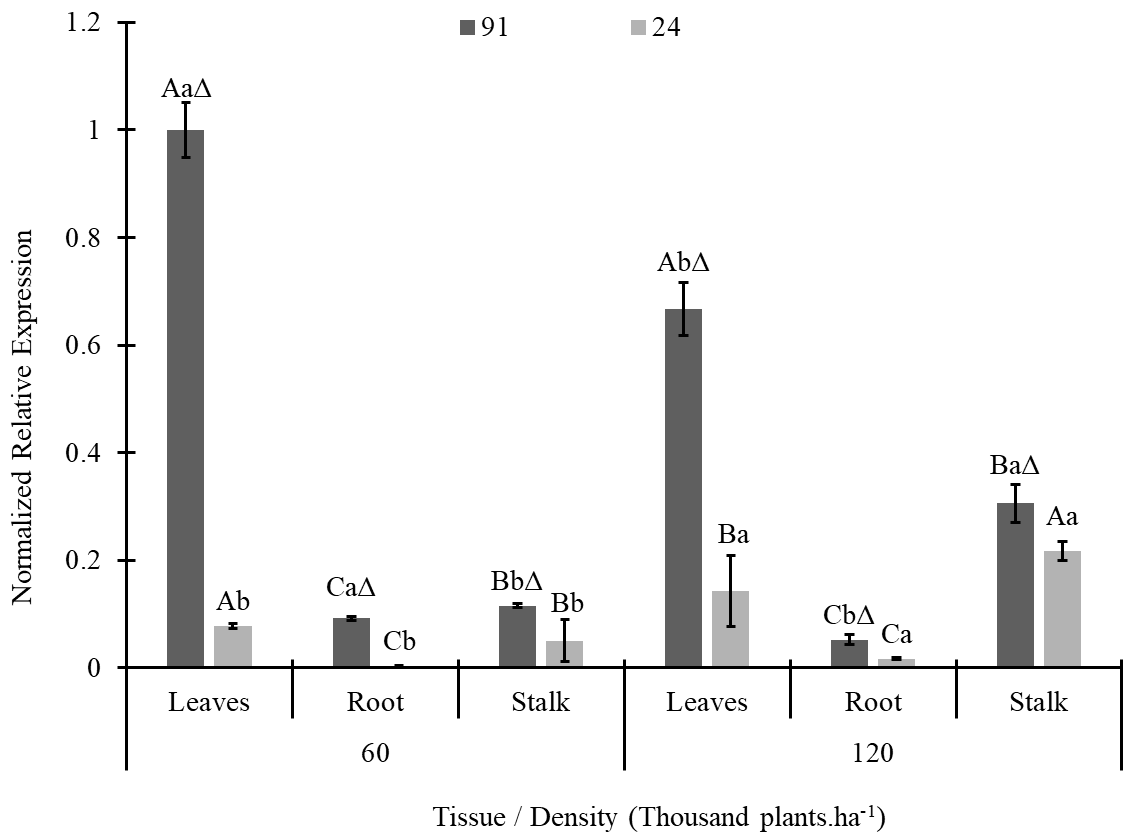
Figure 1. DREB 2A/2.1S gene expression in plant tissues of two maize lines (L91-T and L24-NT) grown at different planting densities (60 and 120 thousand plants.ha-1). The same letters over the bars indicate no differences among treatments in each lineage according to Tukey’s test at 5% probability, with uppercase comparing tissues at each density and lowercase comparing density on each tissue. Sybol Δ indicates that the treatment is different from its equivalent on lineage 24.
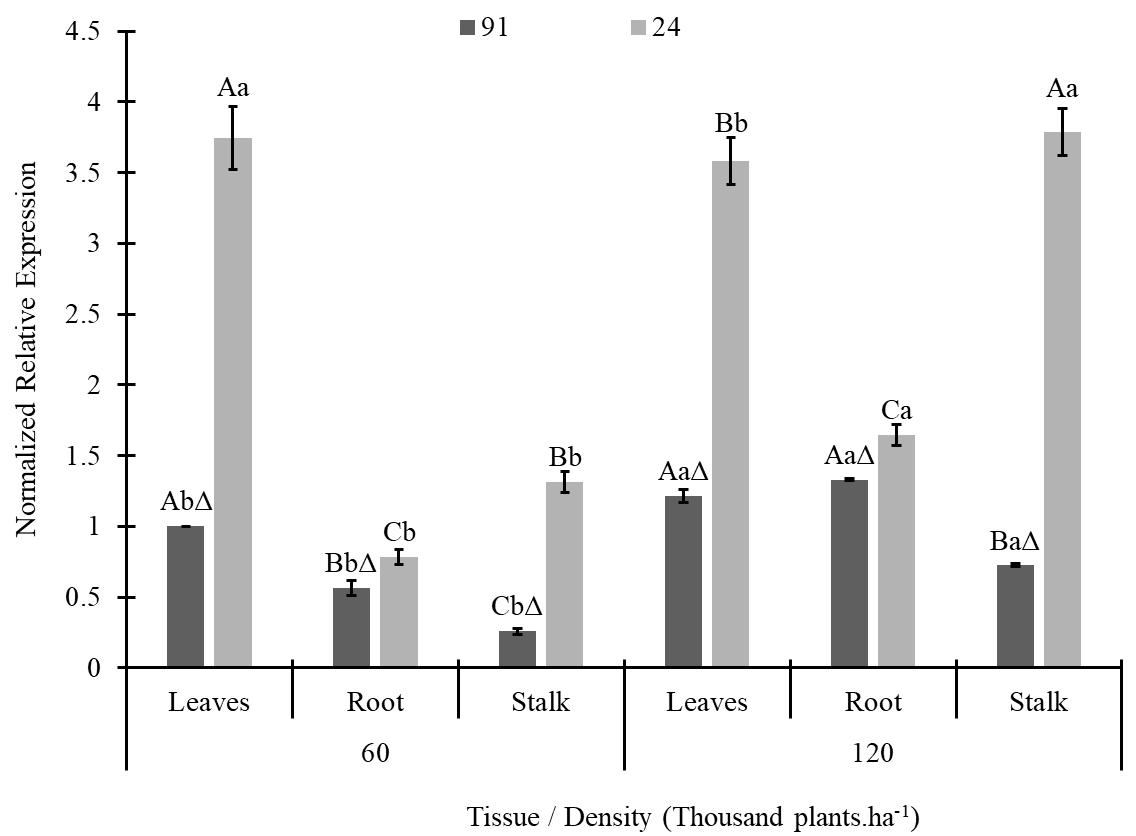
Figure 2. ZmPP2C gene expression in plant tissues of two maize lines (L91T and L24-NT) grown at different planting densities (60 and 120 thousand plants.ha-1). The same letters over the bars indicate no differences among treatments in each lineage according to Tukey’s test at 5% probability, with uppercase comparing tissues at each density and lowercase comparing density on each tissue. Sybol Δ indicates that the treatment is different from its equivalent on lineage 24.
Zhang et al. (2019) reported that a density greater than 60 thousand plants.ha-1 may lead to drought stress due to excessive evapotranspiration (Echarte et al., 2019), with a density of 120 thousand plants.ha-1 being an excessive level, which leads to greater crop stress. In this case, the activation of drought response mechanisms in plants at relatively high densities is expected to occur.
The association between the overexpression of the genes that encode the DREB protein and greater tolerance to water deficit has been reported by several authors and in different crops (Yoshida et al., 2010; Cui et al., 2011). The differential expression of DREB family genes depending on plant tissues, as observed in this research, was also reported for Arabidopsis thaliana and beans (Agarwal et al., 2006). In maize, Santos et al. (2021) reported greater expression of DREB2A/2.1S in aerial plant parts under nonstress conditions, while greater expression was detected in roots under water deficit conditions.
Notably, differences in the expression of DREB family genes may be observed. Although we detected increased expression of the DREB2A/2.1S gene in leaves and in drought-tolerant lines, other studies have shown differential results, as
indicated by the results of Santos et al. (2021) for DREB2A/2.1S and Liu et al. (2013) for ZmDREB, who reported increased expression in roots; additionally, increased expression of this gene was also observed in leaves. These results corroborate those observed for DREB2A/2.1S in our research, which explains why DREB2A/2.1S was more highly expressed in leaves from line 91.
In contrast to DREB2A/2.1S, greater expression of the ZmPP2C gene was detected in the nontolerant line (24). This gene acts in plants as a negative regulator of drought responses; i.e., the increase in its expression can be related to a decrease in water deficit tolerance (He et al., 2019). The overexpression of ZmPP2C-A2 and ZmPP2C-A6 was correlated with low water deficit tolerance by He et al. (2019), who also reported the induction of this gene in aerial parts and roots of maize plants treated with abscisic acid (ABA). Similarly to other authors studying the expression of this gene (Liu et al., 1998; He et al., 2019; Marques et al., 2021), Santos et al. (2021) reported a decrease in the expression of the ZmPP2C gene in maize aerial parts under water restriction conditions.
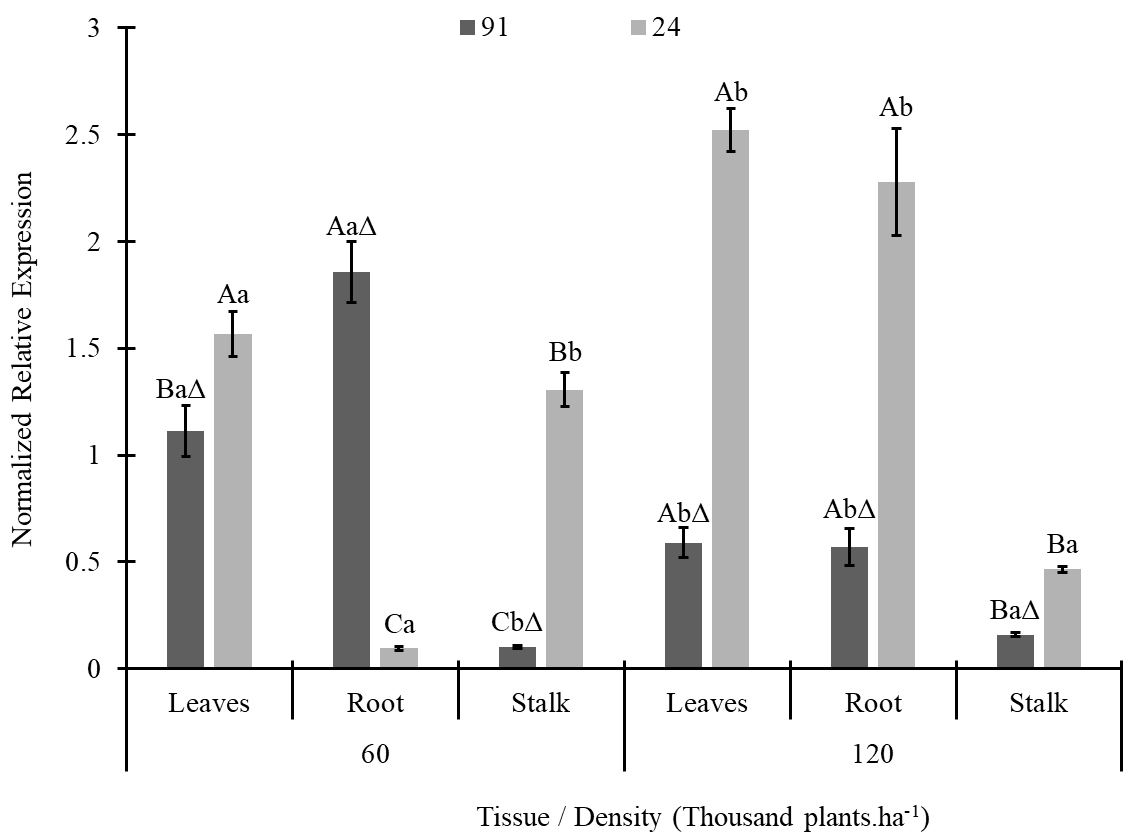
Figure 3. SOD gene expression in plant tissues of two maize lines (L91-T and L24-NT) grown at different planting densities (60 and 120 thousand plants.ha-1). The same letters over the bars indicate no differences among treatments in each lineage according to Tukey’s test at 5% probability, with uppercase comparing tissues at each density and lowercase comparing density on each tissue. Sybol Δ indicates that the treatment is different from its equivalent on lineage 24.
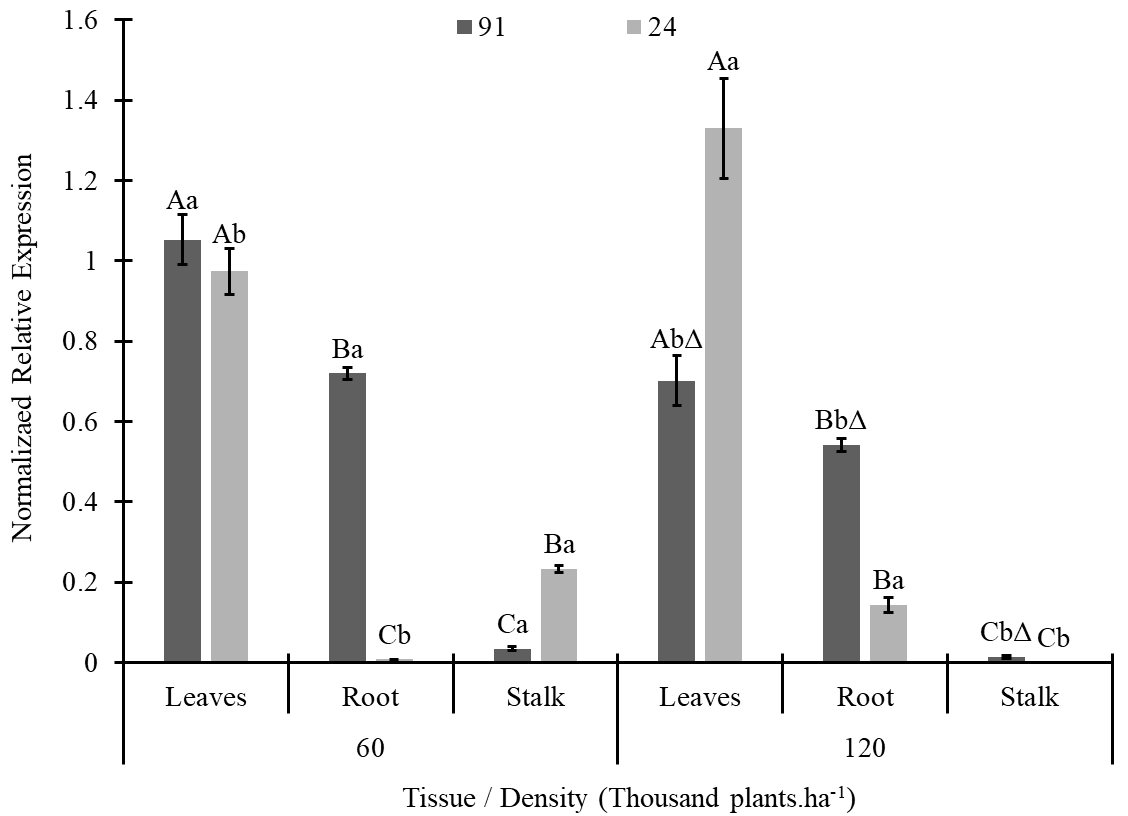
Figure 4. CAT3 gene expression in plant tissues of two maize lines (L91-T and L24-NT) grown at different planting densities (60 and 120 thousand plants.ha-1). The same letters over the bars indicate no differences among treatments in each lineage according to Tukey’s test at 5% probability, with uppercase comparing tissues at each density and lowercase comparing density on each tissue. Sybol Δ indicates that the treatment is different from its equivalent on lineage 24.
ROS accumulation is one of the main causes of stress in plants, and a tolerant line should have an active enzymatic system to avoid this increase in ROS molecules and consequently damage them (Li et al., 2021). Superoxide dismutase (SOD) is the first enzyme to act in the antioxidant system, converting superoxide ions into hydrogen peroxide. Higher levels of SOD in maize under both drought and high temperatures were observed by Hussain (2019), and higher expression of antioxidant system enzymes such as SOD was reported to be correlated with drought-tolerant genotypes (Zhang et al., 2020; Li et al., 2021). However, for our research, higher SOD expression was found in line 24 for all tissues but roots than in line 91 for a density of 120 thousand plants.ha-1, independent of the tissue, and higher expression was observed for the sensitive line.
Following the action of SOD, catalase (CAT) converts H2O2 into water and oxygen, preventing this component from causing further damage to plant cells. The number of genes related to this enzyme has been reported to increase in intolerant maize genotypes under water deficit conditions (Zhang et al., 2020). The expression of the CAT3 gene in our research was greater in line 91, at 60,000 plants.ha-1, which was highlighted in the leaves; in contrast, in line 24, the highest expression was observed at 120,000 plants.ha-1, which was also highlighted in the leaves, where greater values were observed.
For plant breeding programs, faster characterization of the progenies is essential for their selection. The presence of drought tolerance in the progenies is an important trait for maize, and observing agronomical traits in plants under stress conditions may not be an ideal tool because it would require more time, space, and training (Cavasin et al., 2023). On the other hand, many studies have shown the possibility of using alternative techniques for improving progeny selection (Wei et al. 2021; Cavasin et al., 2023). Therefore, the expression of genes related to these characteristics can be an important tool for accelerating this process. However, this requires the study of target gene expression, as conducted in our research, as also shown for lettuce by Cavasin et al. (2023). In our research, we observed clear patterns of expression of DREB2A/2.1S and ZmPP2C in maize, allowing the differentiation of lines 91 (tolerant) and 24 (sensitive) to drought. Based on these results, further studies and adaptations may be conducted to confirm the use of these genes as molecular markers for drought tolerance detection in maize.
Material and Methods
Experimental area
Field experiments were carried out in two experimental areas of the Department of Agriculture of the Federal University of Lavras during two agricultural seasons (2019 and 2020). In 2019, the plant was installed in the experimental area of the municipality of Lavras, MG, whose geographic coordinates are 21°14'S, 40°17'W and 918.80 m. In 2020, the experiment was installed at the Technology Development and Transfer Center (CDTT/DAG/UFLA) of the Federal University of Lavras (UFLA), located in the municipality of Ijaci, MG, whose geographic coordinates are 21°09'S, 44°54'W and 920 m. All evaluations were carried out at the Central Seed Laboratory, located in the Seed Sector of the Department of Agriculture at ESAL/UFLA.
Field conditions
The selection of breeding lines used in the research was carried out according to the results obtained by Abreu et al. (2017), which were based on germination and seedling growth data under conditions of water restriction. Additionally, we used sowing at double the recommended rate for the lines to simulate drought (Monneveux and Ribaut 2006; Sangoi et al. 1996). From the agronomical analysis of maize lines (unpublished data), two strains were selected—L91T (tolerant) and L24-NT (nontolerant). For plant collection, sowing was performed at two densities, one of which is recommended for crops and the other of which may lead to drought stress—60 and 120 thousand plants.ha-1, respectively (Liu et al. 2014; Ren et al., 2014; Zhang et al. 2019). The experimental design used was randomized blocks, with four replications, in a split-plot space, with different plant densities arranged in the plot. Each plot consisted of 4 rows of 16 metres, spaced 0.6 m between rows. The subplots were composed of 4 rows 4 meters in length each. The two central sowing rows of each subplot were considered useful plots. Sowing was performed manually, and after 30 days, thinning was performed to adjust the desired plant population (60 and 120 thousand plants ha-1).
Top dressing fertilizations, as well as other cultural and phytosanitary treatments, were carried out in accordance with the recommendations for the crop. Grain harvesting in both seasons was carried out in the spikes. The ears were harvested and husked manually when the grains reached 20% water content. The ears were dried in small-scale experimental dryers, as described by Navratil and Burris (1982).
Gene expression design and vegetative material collection
The experiment was designed in a completely random design with a triple factorial design of 2 (lines) × 2 (sowing densities) × 3 (tissues). As mentioned above, 91 (tolerant) and 24 (nontolerant) lines were selected. The sowing densities were 60 and 120 plants.ha-1, and the tissues were leaves, stalks and adventitious roots. All the tissues were collected 101 days after sowing from plants at the R1 stage. We collected the youngest and healthiest leaves from each plant; stalks were cut from the penultimate internode, and adventitious roots were those visible immediately above the ground. Immediately after collection, the tissues were frozen in liquid nitrogen, transferred to the laboratory and kept in a deep freezer at -80°C until analysis.
For the gene expression study, we selected genes that were reported to be drought related. Thus, we analysed the expression of DREB2A/2.1S (LOC732788) and ZmPP2C (LOC542176). Additionally, genes related to the synthesis of the enzymes catalase, CAT3 (LOC542370), and superoxide dismutase (LOC100136885) were also used.
RNA extraction and purification
The RT‒qPCR analysis of the transcripts involved in water deficit tolerance was divided into four steps: RNA extraction and purification, reverse transcription for cDNA synthesis, real-time PCR, and analysis of the results. For RNA extraction, the leaves, stems, and adventitious roots of corn lineage plants were macerated in the presence of liquid nitrogen and with the addition of Purification Reagent® (Invitrogen), which was used as an extracting agent.
The samples were macerated in a porcelain crucible with the aid of a pestle, and liquid nitrogen was added to which polyvinylpyrrolidone (PVP) was added to prevent thawing and oxidation of the material. After that, 150 mg of the material was separated into 2 ml microtubes, to which 750 µl of the extracting agent Pure Link Plant Reaget® (Invitrogen) was added. The extraction and purification protocols were carried out according to the manufacturer’s instructions.
The mixture (vegetative material + extracting agent) was vortexed for 2 minutes and kept at rest for 10 minutes under bench conditions, after which the mixture was centrifuged (14,000 rpm/4°C). The supernatant (approx. 600 µL) was collected and subjected to washing, 100 µL of 5 M NaCl was added, and the mixture was stirred for 10 seconds; 300 µL of chloroform was added, and the mixture was stirred again for 1 minute. The tubes were again centrifuged for 10 minutes (14,000 rpm/4°C). An aliquot of 600 µL of supernatant was collected, and the washing procedure was repeated. After centrifugation, 600 µL of supernatant was collected, after which an equal amount of ice-cold isopropanol (approximately 10°C) was added, and the mixture was kept at -20°C for 2 hours. Another centrifugation step was carried out for 25 minutes (14,000 rpm/4°C). The supernatant was discarded, and the pellet was washed by adding 600 µL of 75% ethanol and subsequently centrifuged for 5 minutes (14,000 rpm/4°C). The pellets were dried in a laminar flow hood until the residual ethanol was eliminated, after which the samples were resuspended in 20 µL of ultrapure water.
RNA integrity and purity were evaluated at all stages by means of electrophoresis in a 1.5% agarose gel stained with ethidium bromide and a spectrophotometer (BioTekᵀᴹ Eonᵀᴹ Microplate Spectrophotometer). Electrophoresis was performed for 40 minutes at 80 V and 100 mA, and the gel was visualized in a transilluminator to confirm the integrity of the extracted RNA.
DNAse treatment and cDNA synthesis
After nucleic acid extraction, the samples were DNAse free to avoid any contamination with genomic DNA. For this purpose, the KitDNAse PROMEGA® was used according to the protocol recommended by the manufacturer. For quantification, 5 µg of RNA was added to 2.5 µL of buffer and 0.5 µL of enzyme, and the volume of each sample was adjusted to 25 µL. The mixture was incubated for 30 minutes at 37°C, after which stop reagent was added, and the mixture was kept under bench conditions for 5 minutes to terminate the reaction. The samples were centrifuged (10,000 rpm/4°C) for 5 minutes, after which the supernatant (approximately 20 µL) was collected. To ensure RNA integrity, a second electrophoresis and quantification step was performed with a spectrophotometer (as mentioned above).
cDNA was synthesized from the resulting RNA using the High-Capacity DNA Reverse-to-Transcription cDNA Kit (Applied Biosystems) according to the manufacturer’s protocol. A volume containing 1.5 µg of RNA was collected from each sample, to which 2 µL of buffer, 0.8 µL of nucleotides, 2 µL of random primers and 1 µL of enzyme were added, and the volume was brought to 25 µL with ultrapure water. The material was incubated in a Labnet MultiGene™ thermocycler under the following conditions: annealing at 25°C for 10 minutes, extension at 37°C for 120 minutes, and denaturation at 85°C for 5 minutes. After the reaction, the samples were quantified via a spectrophotometer and diluted 1:5 (sample:water) for RT‒qPCR.
Primer design and efficiency tests
The target and reference genes were selected based on a bibliographic review of studies on water deficit tolerance in maize and other crop species. The sequences of the selected genes used were found in the Maize Genetics and Genomics Database (Woodhouse et al., 2021). Based on these findings, primers were designed using Primer Express 3.0 software (Applied Biosystems). The primers used are shown in Supplementary Table 1. The ubiquitin (UBI) and 18S (18S) genes were used as endogenous controls (Cui et al. 2011; Manoli et al. 2012).
To analyse primer efficiency, 1 µL of each treatment was mixed in a single sample (cDNA pool). The primer efficiency was assessed by applying 1 µL of the cDNA pool to 3 µL of SYBR Green reagent (Applied Biosystems), 0.4 µL of each primer (reverse and forward) and ultrapure water to a final volume of 7 µL. For each primer, six replicates were subjected to RT‒qPCR on an ABI Prism 7500 Real-Time PCR device (Applied Biosystems) via the following steps:
a) Initiation: 2 minutes at 50°C followed by 10 to 95°C.
b) Reaction (45 cycles): 15 seconds at 95°C, 1 minute at 60°C, and 15 minutes at 95°C.
c) Specificity of the reaction according to the denaturation curve (melting) from 60-95°C.
To ensure the absence of contamination from each primer, a blank was prepared by replacing the samples with ultrapure water. The efficiency of the studied primers was determined using LingReg PCR software (Ramakers et al., 2003; Ruijter et al., 2009; Dekkers et al. 2012) following the authors' recommendations. Efficiency was calculated for each of the 4 primers using only primers with an efficiency equal to or greater than 1.8 (equivalent to 90%).
Gene expression assays
Gene expression assays were carried out by using the same mixture mentioned for the primer efficiency assays; only 1 µL of cDNA from each sample was used instead of the cDNA pool. The reaction was performed following the same procedure for primer efficiency. The experiment was carried out with 3 biological replicates, each of which was performed with 3 technical replicates.
Gene expression was analysed using the ∆∆Ct method following the equation used by Hellemans et al. (2008) and using individual efficiency values for each primer, as proposed by Rao et al. (2013). The formulas used to calculate the normalized relative expression of the studied genes are shown in Equation 1. Leaf samples taken after 60 days were used as controls. In this way, the gene expression values were expressed as the normalized relative expression, considering that the expression of the control was equal to 1 and that of the other treatments were expressed as relative values. The relative expression data obtained as described above were analysed through analysis of variance (ANOVA) and Tukey’s test at 5% probability when applicable. All the statistical analyses were carried out by using the software R for Windows (R Core Team, 2022).
Conclusions
Greater expression of the ZmDREB2A/2.1S gene is associated with greater tolerance of corn plants to water deficit. Higher expression of the ZmPP2C gene is associated with lower tolerance of corn plants to water deficit. The genes ZmDREB2A/2.1S and ZmPP2C are candidate genes for the selection of genotypes aimed at tolerance to water deficit. The highest expression of genes related to the water deficit tolerance trait was detected in the leaves.
Acknowledgements
The authors thank the research promotional agencies Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES – Brazil), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG – Brazil), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (Process 426309/2018-9). HOS is a productivity fellow from CNPq (Process 310211/2021-2).
References
Abreu VM, Von Pinho EVDR, Santos HO, Carvalho MR, Naves GMF, Von Pinho, RG (2017) Indirect selection for drought tolerance in maize through agronomic traits and seeds. Rev Bras Milho e Sorgo. 16:287.
Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25:1263–1274.
Al-Kaisi MM, Yin X (2003) Effects of nitrogen rate, irrigation rate, and plant population on corn yield and water use efficiency. Agron J. 95(6): 1475–1482
Benko-Iseppon AM, Soares-Cavalcanti NM, Berlarmino LC, Bezerra Betim JP, Amorin LLB, Ferreira Neto JRC, Pandolfi V, Azevedo HMA, Silva RLO, Santos MG, Alves MVS, Kido, EA (2012) Prospecção de Genes de Resistência à Seca e à Salinidade em Plantas Nativas e Cultivadas. Rev Bras Geogr Física. 4:1112-1134.
Chen J-Q, Meng X-P, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30:2191–2198.
Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J. 90:856–867.
Cui M, Zhang W, Zhang Q, Zhang Q, Xu Z, Zhu Z, Duan F, Wu R (2011) Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol Biochem. 49:1384–1391.
Das K, Roychoudhury A (2014) Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci. 2:1–13.
Dekkers BJW, Willems L, Bassel GW, Bolderen-Veldkamp RP, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT‒qPCR expression analysis in arabidopsis and tomato seeds. Plant Cell Physiol. 53:28–37.
Echarte L, Echarte MM, Cerrudo D, Gonzalez VH, Alfonso C, Cambareri M, Hernandez MD, Nagore ML, Della Maggiora A (2020) Sunflower evapotranspiration and water use efficiency in response to plant density. Crop Sci. 60:357–366.
Fritsche-Neto R, Dovale JC, Cavatte PC (2011) Melhoramento para tolerância a estresses ou para eficiência no uso de recursos. In: Fritsche-Neto R, Borém A (Eds.) Melhoramento de plantas para condições estresses abióticos, 1st edn., Suprema, Visconde do Rio Branco.
He Z, Wu J, Sun X, Dai M (2019) The Maize Clade A PP2C Phosphatases play critical roles in multiple abiotic stress responses. Int J Mol Sci. 20: 3573.
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandersompele J (2008) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:1-14
Hernández MD, Alfonso C, Cerrudo A, Cambareri M, Della Maggiora A, Barbieri P, Echarte MM, Echarte L (2020) Eco-physiological processes underlying maize water use efficiency response to plant density under contrasting water regimes. F Crop Res. 254:107844.
Hussain HA, Men S, Hussain S, Chen Y, Ali S, Zhang S, Zhang K, Li Y, Xu Q, Liao C, Wang L (2019) Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci Rep. 9:3890.
Leng P, Zhao J (2020) Transcription factors as molecular switches to regulate drought adaptation in maize. Theor Appl Genet. 133:1455–1465.
Li H, Yang M, Zhao C, Wang Y, Zhang R (2021) Physiological and proteomic analyses revealed the response mechanisms of two different drought-resistant maize varieties. BMC Plant Biol. 21:1–15.
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 10:1391–1406.
Liu S, Wang X, Wang H, Wang H, Xin H, Yang X, Yan J, Li J, Tran LSP, Shinozaki K, Yamaguschi-Shinozaki K, Quin F (2013) Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9:1-17.
Magaia E, Famba S, Wesström I, Brito R, Joel A (2017) Modelling maize yield response to plant density and water and nitrogen supply in a semiarid region. F Crop Res. 205:170–181.
Manoli A, Sturaro A, Trevisan S, Quaggiotti S (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol. 169:807–815.
Marchin RM, Ossola A, Leishman MR, Ellsworth DS (2020) A Simple Method for Simulating Drought Effects on Plants. Front Plant Sci. 10:1715.
Marques TL, Von Pinho RG, Von Pinho ÉVR, Paniago BC, Freitas NC, Santos HO (2020) Physiological analysis and gene expression analysis of ZmDBP3, ZmALDH9, ZmAN13, and ZmDREB2A in maize lines. Acta Sci – Agron. 42: e43479.
Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, Sasaki R, Suzuki H, Saito K, Shibata D, Shinozaki K, Yamagushi-Shinozaki K (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150:1972–1980.
Monneveux P, Ribaut JM (2006) Secondary traits for drought tolerance improvement in cereals. In: Ribaut JM (Ed) Drought adaptation in cereals. CABI Direct, Binghamton.
Navratil RJ, Burris JS (1982) Small‐Scale Dryer Design. Agron J. 74:159–161.
Nepomuceno AL, Stewart JMCD, Oosterhuis D, Turley R, Neumaier M, Farias JRB (2000) Isolation of a cotton NADP (H) oxidase homologue induced by drought stress. Pesqui Agropecuária Bras. 35:1407.
Oliveira DF, Cavasin PY, Silva S, Oliveira NS, Oliveira CL, Gomes LAA (2021) Genetic control of thermoinhibition tolerance in lettuce seeds. Pesqui Agropecuária Bras. 56:e02337.
Pérez-Alfocea F, Ghanem ME, Gómez-Cadenas A, Dodd IC (2011) Omics of Root-to-Shoot Signalling Under Salt Stress and Water Deficit. Omi A J Integr Biol. 15:893–901.
R Core Team (2022) R: A Language and Environment for Statistical Computing
Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 339:62–66.
Rao X, Huang X, Zhou Z, Lin X (2013) An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 3:71–85.
Ren X, Sun D, Wang Q (2016) Modelling the effects of plant density on maize productivity and water balance in the Loess Plateau of China. Agric Water Manag. 171:40–48.
Roy SJ, Tucker EJ, Tester M (2011) Genetic analysis of abiotic stress tolerance in crops. Curr Opin Plant Biol 14:232–239.
Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45–e45.
Santos MC, Von PINHO ÉV de R, Dos Santos HO, Vilela DR, Silva Neta IC, Abreu VM, Vasconcellos RCC (2021) Enzymatic activity and gene expression related to drought stress tolerance in maize seeds and seedlings. Biosci J. 37:e37079.
Sani BM, Oluwasemire KO, Mohammed HI (2008) Effect of irrigation and plant density on the growth, yield and water use efficiency of early maize in the nigerian savanna. J Agric Biol Sci. 3:121-131.
Sangoi L, Salvador R (1996) Agronomic performance of male-sterile and fertile maize genotypes at two plant populations. Ciência Rural. 26:377–383.
Verma AK (2016) Abiotic Stress and Crop Improvement: Current Scenario. Adv Plants Agric Res. 4:346-346.
Wei X, Fan X, Zhang H, Jiao P, Jiang Z, Lu X, Liu S, Guan S, Ma Y (2022) Overexpression of ZmSRG7 improves drought and salt tolerance in maize (Zea mays L.). Int J Mol Sci. 23:13349.
Winans ET, Beyrer TA, Below FE (2021) Managing Density Stress to Close the Maize Yield Gap. Front Plant Sci 12: 767465.
Woodhouse MR, Cannon EK, Portwood JL, Harper LC, Gardiner JM, Schaeffer ML, Andorf CM. (2021) A pangenomic approach to genome databases using maize as a model system. BMC Plant Biol 21: 385.
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signalling involved in drought stress tolerance and require ABA for full activation. Plant J. 61:672–685.
Zhang A, Jiang M, Zhang J, Tan M, Hu X (2006) Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141:475–487.
Zhang Y, Wang R, Wang S, et al. (2019) Effect of planting density on deep soil water and maize yield on the Loess Plateau of China. Agric Water Manag. 223:105655.
Zhang Q, Liu H, Wu X, Wang W (2020) Identification of drought tolerant mechanisms in a drought-tolerant maize mutant based on physiological, biochemical and transcriptomic analyses. BMC Plant Biol. 20:315.