

Aust J Crop Sci. 18(11):760-767 (2024) | ISSN:1835-2707
https://doi.org/10.21475/ajcs.24.18.11.p159
Comparative study of two annual Medicago species: Cytogenetical approaches and seed storage proteins analysis in M. polymorpha L. and M. laciniata (L.) Mill.
Lacheheb Fairouz* and Belkhodja Moulay
Laboratory of Experimental Biotoxicologie, Bioremediation and Phytoremediation
University of Oran 1. Ahmed Benbella, BP 1524 ELM_Naouer 31000, Oran. Algeria
*Corresponding author: Lacheheb Fairouz  and Belkhodja
Moulay
and Belkhodja
Moulay 
Abstract: Species of the Medicago genus are of great interest for their ability to fix atmospheric nitrogen. Only a few species, mainly those used in agriculture, have been well studied. There is a particular lack of study of the annual wild species, which are represented in very small numbers (4/103) in the genus. This is why we have chosen to study two annual species with two different basic chromosome numbers, Medicago laciniata (L.) Mill. x = 8 and Medicago polymorpha L. x = 7. The main questions were to understand how the decrease in basic chromosome number from x=8 to x=7 occurred and what is the relationship between these species. To answer these questions, the karyotype was analysed using conventional karyological methods and the genome size was assessed using flow cytometry. Ten accessions of M. polymorpha and six of M. Laciniata were studied for cytogenetical feature (by classical karyological methods). Apart from the basic chromosome number, some differences in karyotype features such as chromosome size and morphology were observed. The mean genome size values were 2C = 1.817 pg for M. Laciniata and2C=1.123 pg for M. polymorpha. The SDS-PAGE gel analysis of the seed storage proteins showed species-specific polypeptides pattern. In addition to differences in the basic chromosomes number, karyotype characteristics and total nuclear DNA content, protein profiles also differ between the two species. We suggest that decrease in basic chromosome number from x=8 to x=7 is probably due to occurrence of Robertsonian fusion or centric fusion.
Keywords: Annual Medicago species, flow cytometry, karyotype, Leptospirae section, total proteins.
Abbreviations: C_ Total amount of DNA contained in a haploid nucleus; CT_ chromosome type; M_Medicago; m_metacentric; pg_picogramme SC_secondary constriction; Sm_Submetacentric.
Introduction
Like all legumes, species of Medicago genus have an important agro-economic interest, which lies in their ability to fix atmospheric nitrogen, enabling soil fertilization and limiting the input of fertilizers in a rotation system with other plants (Abdelguerfi et al., 1988; Small, 2011; Yahia and Fyad-Lamèche, 2003).
Medicago polymorpha is native to Eurasia and Africa, but has been widely introduced elsewhere, as it is not limited by cold, drought and waterlogging (Small and Jomphe, 1989). Moreover, this species is the most ubiquitous and morphologically polymorphic. It grows on acid soils, in the low and salted meadows of the Mediterranean Sea edge or in the more arid scrublands (Prosperiet al., 1995; Prosperietal., 2000). According to Chen et al. (2021), M. polymorpha has been escaped from roadsides, fields, stream banks and has adapted to variable environments. Badri et al. (2016) consider that the hardness and dormancy of M. polymorpha seed, coupled with a highly variable flowering time, have allowed this species to survive unfavourable periods in a wide variety of bioclimatic zones. In Algeria, this species is also adapted to different soil pH (Abdelguerfiet al., 1988).
The natural habitat of Medicago laciniata is in dry, sandy or stony desert-like environments, where it is often the only surviving species of the genus Medicago. The species appears to be native to countries along the southern coast of the Mediterranean Sea (Lesins and Lesins, 1979). Small and Jomphe (1989) also indicate that the M. laciniata is probably native to southern and eastern Mediterranean countries where it appears to be adapted to steppe and desert soils but largely introduced as a ruderal weed. In Algeria, this species grows at low altitudes in soil with medium-sized particles and prefers a slightly alkaline to alkaline pH (Abdelguerfiet al., 1988).
Cytogenetic studies of Medicago species are quite numerous (Small and Bauchan, 1984; Quiros and Bauchan, 1988; Mariani et al., 1996; Cerbah et al., 1999; Sadeghian and Hejazi, 2014) but they are scarce for M. polymorpha and especially for M. laciniata (Heyn, 1963; Mariani et al., 1996; Sadeghian and Hejazi, 2014; Djafri-Bouallag et al., 2019).
Over 95% of the species in the genus Medicago have a basic chromosome number x=8, and only four species have x=7, including M. polymorpha, M. Constricta Dur, M. praecox DC and M. Rigidula L (Falistocco, 2018). In their phylogenetic study based on sequencing of the TRN/MATK and GA3ox1 genes, Steele et al. (2010) highlighted that in the Medicago genus, the reduction from (2n=16 to 2n=14) has most likely occurred at least three times.
Three levels of ploidy were detected in the genus: diploid (2n=2x=14 and 2n=2x=16) in the majority of annual and perennial species, tetraploid (2n=4x=32) in perennial species and two annuals [M. scutellata (L.) Milland M. rugosa Desr.] and hexaploid (2n=6x=48) in only a few perennials species as M. Arborea L., M. Saxatilis M. Bieb., M. Cancellata M. Bieb. (Lesins and Lesins,1979; Quiros and Bauchan, 1988; Small and Jomph, 1989). All species of Leptospirae section have 2n=16 except M. polymorpha and M. Praecox DC. which have (2n=14).
In general, the seed storage protein profile on SDS-PAGE can be used for varietal identification and biosystematic studies between different species (Krochko and Bewley,1988; Fyad-Lamèche,1998; Yin et al.,1998; Krochko and Bewley, 2000; Panigrahi et al., 2007; Singh et al., 2018; Rajpal et al., 2022).
The seed proteins are mainly reserve proteins and do not undergo changes in the mature seeds. Their composition is very stable and almost unaffected by environmental conditions or seasonal fluctuations (Ladizinsky and Hymowitz, 1979). Thus, seed storage proteins generally represent a large number of stable genetic markers. Electrophoretic analysis of these proteins has the advantage over traditional identification techniques, offering a rapid and reliable assessment of genotypic differences between most cultivars of Phleum pratense L. (Cai and Bullen, 1992). Other studies have shown that intrinsic changes in the cell, such as chromosomal rearrangements or even change in chromosome number, have no effect on the protein profile (Ladizinsky and Hymowitz, 1979).
The aims of the present study were: (1) to verify the chromosome number, characterize the karyotypes and establish the idiograms of M. polymorpha and M. Laciniata using classical karyological method; (2) to estimate the nuclear DNA content by flow cytometry and (3) to analyse the electrophoresis of seed storage proteins. All these characteristics could be useful for a better understanding of the relationship between the two species studied. In addition, the importance of the seed storage protein profile as a species-specific trait will be discussed.
Results
Chromosome number and karyotype feature
The chromosome number was diploid in both species, 2n=2x=14 for M. polymorpha, and 2n=2x=16 for M. Laciniata), indicating a different basic chromosome number of x=7 and x=8, respectively.
In addition, the two species showed different karyotype features that were revealed by morphometric analysis (Table 1a, 1b). After Feulgen staining, the karyotypes of M. Laciniata (Figure. 1a and b) and that of M. Polymorpha (Figure. 1c and d) presented differences in the type of chromosomes and position of secondary constrictions (SC). In M. laciniata the SC was found on one of the large chromosome pairs, whereas in M. polymorpha it was on one of the small chromosome pairs (Figure. 1 c and d, arrows).
Regarding the inter-chromosomic asymmetry, which is due to the heterogeneity of chromosomal sizes, we estimated the coefficient of chromosome length variation (CVCL). The CVCL relative dispersion parameter was 0.30 for M. polymorpha and 0.15 for M. Laciniata (Table 2) which has a more symmetrical karyotype than M. polymorpha. The asymmetry linked to chromosome size was also assessed by the R value (ratio between the longest and the shortest pair), which was 1.38 in M. Laciniata and 2.31 in M. Polymorpha (Table3).
The average length of chromosomes varies from 1.26 µm to 2.9 µm in M. polymorpha and from 1.62 µm to 2.42 µm for M. laciniata. The total length of the haploid karyotype was 13.035 µm in M. Polymorpha and15.696 µm in M. Laciniata (Table 2a and b). The relative length of chromosomes of each species is also shown in Table 2a and b. For the chromosome pair 1, the total relative length of 22.252 for M. polymorpha differed considerably from that of 15.401 for M. laciniata.
The karyograms of both species show the presence of two chromosome types, metacentric (m) and submetacentric (sm). The karyotype formula was 2n=2x=14=12m+2sm for M. polymorpha and 2n=2x=16=10m+6sm for M. laciniata. The karyotype formula shows relatively symmetrical karyotypes in both species and the asymmetry index was 58.898 for M. polymorpha and 63.163% for M. laciniata (Table 3).
The calculation of "A" parameter according to Watanabe et al. (1999) for the same purpose gave two significantly different values, 0.167 for M. polymorpha and 0.257 for M. Laciniata (Table 4). Finally, three pairs of chromosomes have been determined easily and with certainty in M. polymorpha, whereas the longest and the shortest pairs and also the submetacentric pair bearing secondary constriction. The chromosomes of M. laciniata are very similar in morphology and total length. However, two homogeneous groups of chromosomes seem to characterize this karyotype; two (or three) large and five small pairs of chromosomes. Only the first two pairs, the longest in the chromosome set, and the last pair carrying the secondary constriction, could be identified (Figure 1).
Genome size evaluation
The ten accessions of the M. polymorpha were measured for genome size evaluation. The 2C DNA values ranged from 1.11 to 1.148 pg, the mean value was 1.123 pg. In the three M. laciniata accessions the values ranged from 1.804 to 1.834 and the mean value was of (1.817pg) (Table 5; Figure 2a and b). Differences in 2C-values among accessions of both species are not significant despite the diverse geographical origins and environmental conditions of these accessions.
The two investigated species differed significantly from each other in terms of DNA content (Figure 2c) which was expected considering their different basic chromosome numbers and chromosome sizes.
Seeds storage protein analysis
Three different areas were labelled on gel (Figure 3); cathodic zone (Cz), an intermediate zone (Iz) and an anodic one (Az). Each area contains major and minor bands, according to their molecular weight which ranges from 10 to 230 kDa. In addition, the major bands of molecular weight between 40 and 80 kDa observed on seed storage SDS-PAGE profiles clearly characterize accessions and species.
There are many common bands among accessions of both species. For the accession 21B of M. polymorpha (from Algeria) the largest number (20) of bands with high intensity profiles was observed in column number7 (Figure 3). In the case of M. laciniata, the protein profiles have slightly fewer and less intense bands. Two groups of major bands (high intensity) of molecular weight between 40 and 80 kDa in the intermediate and the cathode zone of the gel are specific for M. polymorpha and are present in the profiles of all accessions of this species. The profiles of M. laciniata accessions also show specific major bands in these two zones. However, they differ completely from those of M. polymorpha. Anodic zone bands are less species-specific (Figure 3).
Discussion
The number of chromosomes and karyotype features vary between M. polymorpha and M. laciniata. The most frequent basic chromosome number in the genus Medicago is x=8. Only few species possess x=7, the number which probably arises during decreasing dysploidy process. For example, in M. murex Willd., it appears that 2n= 14 is derived from 2n = 16 by transferring almost all of the chromosomal material from the 8th (shortest) chromosome pair to the 3rd pair, making it the longest in the x=7 chromosome complement (Lesins et al., 1970; Gillies, 1971; Lesins and Gillies, 1972). This phenomenon has been arisen three times within the genus
Table 1a. Mean morphometric data of the M. Laciniata karyotype.
| N° pair | SA µm ± σ | LA µm ± σ | TL µm ± σ | RTL | r | AsI | R | CT |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.784±0.18 | 1.633±0.30 | 2.417±0.48 | 15.401 | 2.083 | sm | ||
| 2 | 0.792±0.18 | 1.535±0.37 | 2.319±0.42 | 14.776 | 1.938 | sm | ||
| 3 | 0.735±0.10 | 1.307±0.15 | 2.042±0.20 | 13.007 | 1.778 | sm | ||
| 4 | 0.719±0.07 | 1.176±0.21 | 1.895±0.28 | 12.071 | 1.636 | 63.163 | 1.382 | m |
| 5 | 0.739±0.08 | 1.127±0.31 | 1.878±0.36 | 11.967 | 1.525 | m | ||
| 6 | 0.735±0.05 | 1.143±0.20 | 1.878±0.19 | 11.967 | 1.555 | m | ||
| 7 | 0.653±0.07 | 0.996±0.07 | 1.650±0.15 | 10.510 | 1.525 | m | ||
| 8 | 0.621±0.15 | 0.996±0.10 | 1.617±0.08 | 10.302 | 1.604 | m -CS | ||
| Σ=5.778 | Σ = 9.914 | Σ = 15.696 |
SA - Short arm; LA - Long arm; TL - Total length; RTL - Total length on sum of total length (TL / ΣTL) x 100; r - LA / SA; CT - Chromosome-type; AsI - Asymmetry index (Σ LA / Σ TL) x 100; R - Ratio between the length of the largest and smallest pair; sm: Sub metacentric; m : metacentric ; m-CS : metacentric with secondary constriction
Table 1b. Mean morphometric data of the M. Polymorpha karyotype.
| N° pair | SA µm ± σ | LA µm ± σ | TL µm ± σ | RTL | r | AsI | R | CT |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.035±0.310 | 1.847±0.344 | 2.901±0.001 | 22.252 | 1.785 | sm | ||
| 2 | 0.890±0.272 | 1.354±0.186 | 2.244±0.417 | 17.217 | 1.521 | m-CS | ||
| 3 | 0.886±0.287 | 1.053±0.150 | 1.938±0.321 | 14.871 | 1.189 | m | ||
| 4 | 0.686±0.201 | 0.979±0.086 | 1.665±0.171 | 12.775 | 1.426 | 58.898 | 2.307 | m |
| 5 | 0.654±0.178 | 0.947±0.047 | 1.601±0.164 | 12.281 | 1.447 | m | ||
| 6 | 0.677±0.093 | 0.751±0.153 | 1.428±0.219 | 10.958 | 1.110 | m | ||
| 7 | 0.510±0.016 | 0.747±0.061 | 1.257±0.081 | 9.646 | 1.465 | m | ||
| Σ= 5.339 | Σ= 7.677 | Σ= 13.035 |
SA - Short arm; LA - Long arm; TL - Total length; RTL - Total length on sum of total length (TL / ΣTL) x 100; r - LA / SA; CT - Chromosome-type; AsI - Asymmetry index (Σ LA / Σ TL) x 100; R - Ratio between the length of the largest and smallest pair.sm: Sub metacentric; m : Metacentric; m-CS : metacentric with secondary constriction
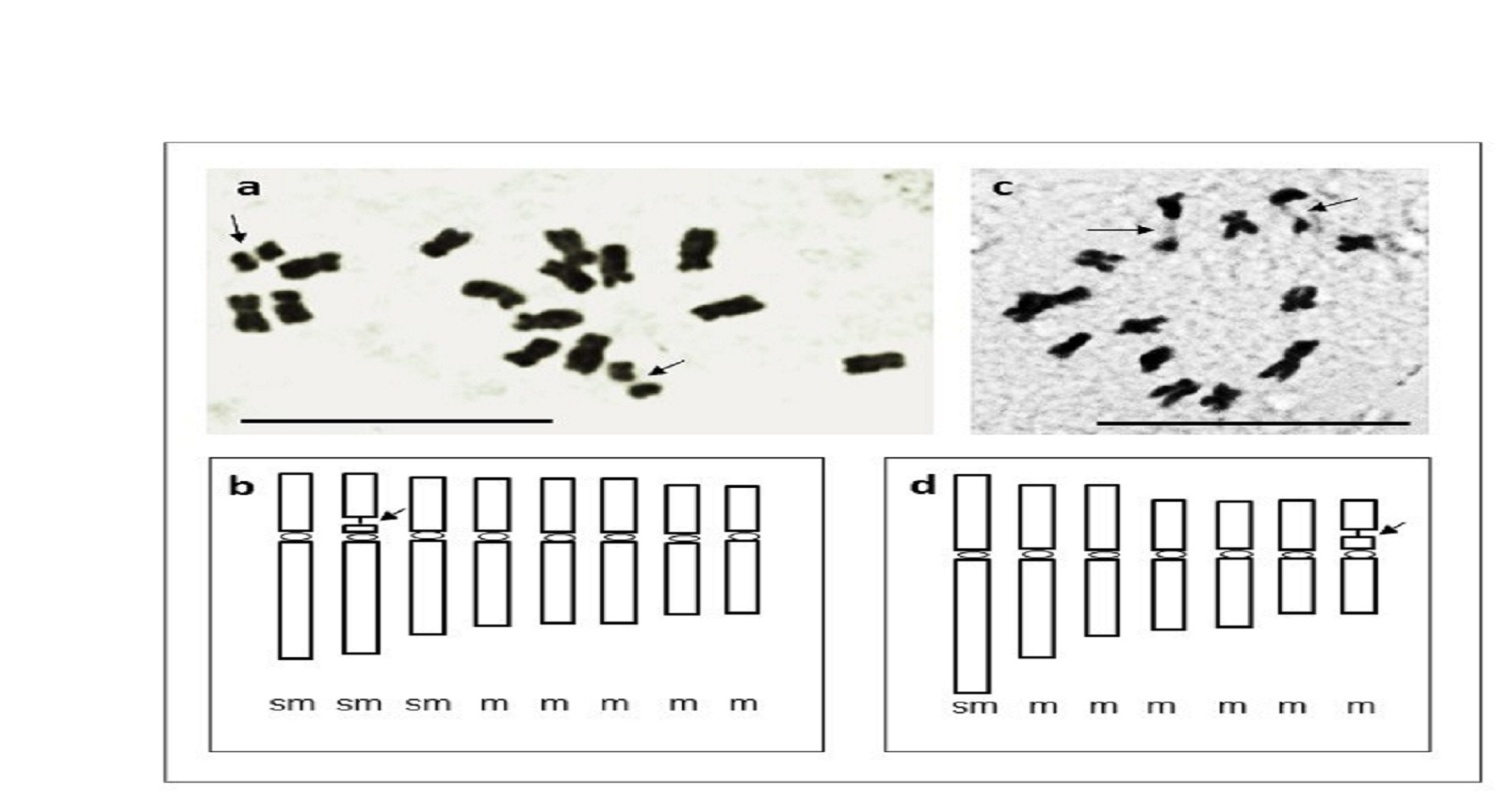
Figure 1. The Feulgen method is utilized for counting chromosomes and studying chromosome types in the two species studied.
(a): Medicago laciniata, 2n=16: Feulgen staining of metaphase chromosome plate. (b): corresponding idiogram with indication of chromosome types and position of secondary constrictions (c): M. polymorpha2n=14: Feulgen staining of metaphase chromosome plate, (d): The diagram represents the secondary constrictions, and the arrow indicates the size of this secondary constriction measured by the unit, a Barr = 10 µm.
Medicago, in Pachyspireae, Leptospireaeb and Geocarpae sections (Steele, 2010). Medicago polymorpha is one of these species with x=7. Some authors report also the observation of 2n =16 chromosomes in this species. We did not observe such chromosome number in our M. polymorpha sample, as also reported by Sadeghian and Hejazi (2014). Mariani et al. (1996) suggested that the two cytotypes (2n=14 and 16) can coexist in M. polymorpha, which could explain the high morphological variability in this species. The 2n=16 has already been reported by Heyn (1963) and Small and Jomphe (1989). However, Lesins and Lesins, (1979) considered that the chromosome number of 2n=16 in M. polymorpha was an erroneous count due to two easily detached satellites. Sadeghian and Hejazi (2014) also considered that the 2n = 16 for M. polymorpha is either due to confusion with other Medicago species or probably overestimation due to the presence of large detached satellites. In our opinion, these are not the satellites but the larger chromosomes' fragments detached at the secondary constriction level which is the most fragile site of chromosome as showed on Figure 1c
(arrows). The fragility of secondary constrictions and/or centromeres, especially when they are in an intercalary position on chromosome, is often the cause of chromosome miscount in plants (Hidalgo et al., 2007; Benmiloud-Mahieddine et al., 2011; Garnatje et al., 2004).
This phenomenon was also observed in Medicago truncatula Gaertn. (Cerbah et al., 1999). Similarly, Bauchan and Elgin, (1984) reported a case of the satellites in Medicago rugosa Desr., which “are large and they are located a relatively long distance from the main body of the chromosome, possibly leading the erroneous previously recorded chromosome count of 2n= 32”.
The two studied species here differed considerably from each other in all morphometric karyotype parameters (long arm, short arm, total length, inter and intra asymmetry indexes). The average values for short arms, long arms and the total length of chromosomes are similar to those reported in the literature for the two species but the karyotype formula differs (Mariani et al.,1996; Sadeghian and Hejazi, 2014). Sadeghian and Hejazi (2014) reported only one chromosome
Table 2. Coefficient of variation for chromosome length of the two investigated species.
| M. laciniata | Pair | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | m | σ | CVCL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TL | 2.42 | 2.32 | 2.04 | 1.89 | 1.88 | 1.88 | 1.65 | 1.62 | 1.96 | 0.29 | 0.148 | |
| M. polymorpha | Pair | 1 | 2 | 3 | 4 | 5 | 6 | 7 | - | |||
| TL | 2.9 | 2.24 | 1.94 | 1.66 | 1.6 | 1.43 | 1.26 | 1.861 | 0.560 | 0.301 |
Tl: total length; m: mean; σ: standard deviation; CT for M. laciniata (Pair: 1,2,3: sm; pair: 4, 5, 5,6: m and pair: 8: m-CS). CT for M. polymorpha (Pair: 1: sm; pair: 3,4,5,6,7: m and pair: 2: m-CS).
Table 3. Synthesisof karyotype data for two studied species
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
x: basic chromosome numbers; Ʃ TL: total length of the chromosome; As I %: asymmetry index (Ʃ long arms / Ʃ short arms x 100); R: Ratio between the longest and shortest pair.
Table 4. The parameters that determine the size of chromosome pairs.
| Chromosome pair | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| M. laciniata | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | mean |
| A = (L –S)/(L+S) | 0.351 | 0.319 | 0.28 | 0.241 | 0.208 | 0.217 | 2.08 | 0.232 | 0.257 |
| M. polymorpha | 1 | 2 | 3 | 4 | 5 | 6 | 7 | - | mean |
| A = (L –S)/(L+S) | 0.28 | 0.205 | 0.087 | 0.174 | 0.183 | 0.053 | 0.188 | - | 0.167 |
L: long arm length; S: short arm length; CT for M. laciniata (Pair: 1,2,3: sm; pair: 4, 5, 5,6: m and pair: 8: m-CS). CT for M. polymorpha (Pair: 1: sm; pair: 3,4,5,6,7: m and pair: 2: m-CS).
type (metacentric) in karyotype of both species, whereas our results show the existence of metacentric and submetacentric pairs in both M. polymorpha and M. laciniata.
Sadeghian and Hejazi (2014) also considered that the 2n=16 for M. polymorpha is either due to confusion with other Medicago species or probably overestimated due to the presence of large detached satellites.
The lower value of the asymmetry index in M. polymorpha may reflect an acquired symmetry i.e. secondary symmetry (Siljak-Yakovlev, 1996) of the karyotype, resulting from rearrangements during the reduction of the chromosome number (the larger chromosome pair is probably the result of Robertsonian fusion or centric fusion).
As a consequence of dysploidy events, which may have led to the reduction of the basic chromosome number from x = 8 to x = 7, all Medicago species with 2n = 14 would descend from a progenitor with 2n = 16 (Falistocco, 2018). To illustrate this fact, this author mentioned the case of M. lesinsii Small. E (2011) and M. murex. Lesinset al. (1970), comparatively analysed pachytene chromosomes, and suggested that M. murex (2n = 14) very likely derived from M. lesinsii (2n = 16) by translocations which results in the formation of a large chromosome and the loss of a centromere.
The relative karyotype uniformity, almost only one chromosome type (metacentric) and one pair of chromosomes carrying a secondary constriction have been pointed out in the bibliography as the general feature of M. polymorpha karyotype (Agarwal and Gupta, 1983; Mariani et al., 1996; Sadeghian and Hejazi, 2014; Falistocco, 2018).
Our results indicate that in M. polymorpha, three pairs of chromosomes were reliably and easily determined: the longest and the shortest ones and also the submetacentric pair bearing secondary constriction. These three pairs of chromosomes could be considered markers in the karyotype of M. polymorpha.
In M. laciniata, several pairs have similar morphology and total length. The two longest pairs are very close in both size and morphology and the pairs 4,5,6 and 7 are also very similar to one another. Only the first two pairs, the longest in the chromosome set, and the last pair which carries the secondary constriction, could be identified. In our accession, the karyotype of M. laciniata showed three submetacentric pairs, whereas Sadeghian and Hejazi, (2014) reported only metacentric chromosome pairs for this species.
The significant difference in DNA content was detected between the two species. As expected, M. laciniata presented a higher 2C value than M. polymorpha. However, at the intraspecific level, there is no significant differences among accessions of both species.
It appears that the change in chromosome number from 2n=16 to 2n=14 and the difference in chromosome size may be related to a significant difference in DNA content between the two species.
According to Laamari et al. (2016), variations among populations of M. sativa L. Gabsias were observed, which in some cases could be related to environmental gradients or growing conditions. Kalendar et al. (2000) observed variations in the number of copies of the retrotransposon BARE-1 in wild barley (Hordeum spontaneum L.) populations in response to different microclimates. Furthermore, in Vicia faba L., Ceccarelli et al. (1995) indicated that Fok1 (60 bp tandem repeat) is involved in a part of variation in DNA content between accessions (up to 35% variation) and the copy number of Fok1 increases from 5.4 to 21.5 106 representing up to 9.5% of the genome.
The comparison of the current results and those obtained in our previous study (Fyad-Lamèche et al., 2016) showed the stability of genome size values. In both studies, each species presented a stable and specific genome size value, with a significant interspecies difference and a low-amplitude of intraspecific variation.
Since the genome size is not a random variable, but a constant per species and per accession, any recorded variation must be taken into account. If the technical process of evaluating the genome size cannot be incriminated, the observed intraspecific variation could be due to an effective variation of the genetic material itself, i.e. to a "positive" or "negative" variation in the number of base pairs. All stable, reproducible differences must be taken into account, regardless of their amplitude.
The genome size values obtained in our previous and present studies of M. polymorpha, indicated the existence of only one cytotype (2n=14) among the all investigated accessions. If the two cytotypes (2n=16 and 2n=14) coexisted in the studied accessions, we should find two significantly different values of genome size, one value for each cytotype, which was not the case in our sample. Thus, the results of the cytometric analysis confirmed the cytogenetic results.
Table 5. Genome size of the ten M. polymorpha and three M. laciniata accessions.
| Species | Taxon | Accession No. | 2n | 2C DNA (pg)* |
SD | 1Cx** (Mbp) |
|---|---|---|---|---|---|---|
| M. polymorpha | M. polymorphabrevispina | IFMA 3032 | 14 | 1.118 | 0.013 | 547 |
| M. polymorphavulgaris | IFMA 3907 | 14 | 1.117 | 0.001 | 546 | |
| M. polymorphavulgaris | IFMA 3917 | 14 | 1.128 | 0.009 | 551 | |
| M. polymorphas.l. | IFMA 3930 | 14 | 1.131 | 0.012 | 553 | |
| M. polymorphavulgaris | IFMA 3938 | 14 | 1.148 | 0.022 | 561 | |
| M. polymorphapolymorpha | IFMA 727 | 14 | 1.115 | 0.017 | 545 | |
| M. polymorphas.l. | Site 21B | 14 | 1.124 | 0.006 | 550 | |
| M. polymorphas.l. | Site 214 | 14 | 1.110 | 0.017 | 543 | |
| M. polymorphas.l. | Site 213 | 14 | 1.124 | 0.015 | 550 | |
| M. polymorphas.l. | Site 57 | 14 | 1.117 | 0.017 | 546 | |
| Mean | 1.123 | |||||
| M. laciniata | M. laciniatalaciniata | IFMA 0730 | 16 | 1.834 | - | 896 |
| M. laciniatas.l. | IFMA 2971 | 16 | 1.814 | - | 887 | |
| M. laciniatas.l. | IFMA 3020 | 16 | 1.804 | - | 882 | |
| Mean | 1.817 |
*1pg = 978 Mbp;**1Cx -Monoploid genome size.
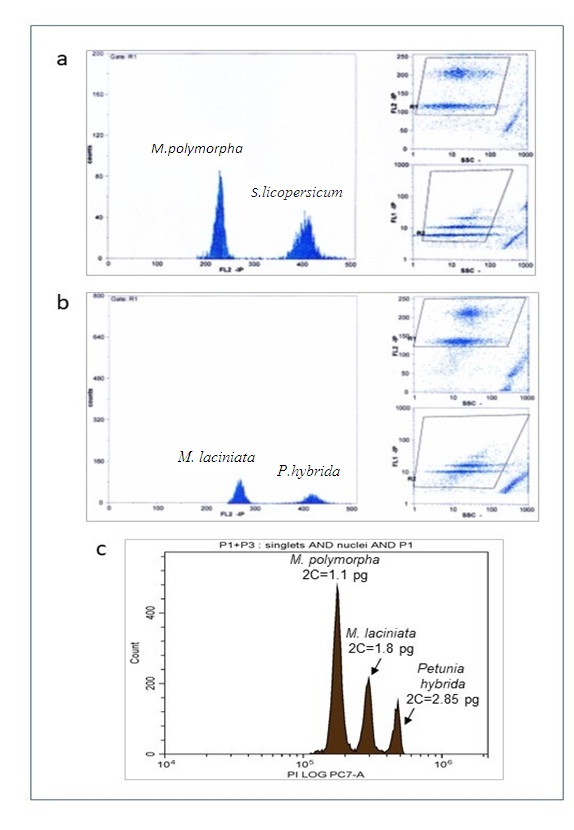
Figure 2. Cytometric histogram of propidium iodide-stained nuclei of cells. (a): The two peaks of M. polymorpha and Solanum licopersicum that are used as a standard are depicted in Figure a. (b): Figure b shows the two peaks of M. laciniata and Petunia. hybrida that are utilized as standards. (c): The standard used for measuring both Medicago species was Petunia hybrida because the images of Solanum licopersicum and M. polymorphaoverlapped.
Seed protein markers are quite stable, uniform, reliable, reproducible and seldom affected by environmental fluctuations and management practices. The seed storage proteins SDS-PAGE analysis is a powerful tool to solve taxonomic or evolutionary problems (Panigrahi et al., 2007). These species-specific polypeptides can also be used as markers for species identification as well as for hybrid characterization in an introgressive hybridization programmes (Panigrahi et al., 2007).
In this study, nine accessions of M. polymorpha and five of M. laciniata were analysed by SDS-PAGE. The profiles obtained allow M. polymorpha accessions to be easily distinguished from M. laciniata accessions. Intraspecific variability exists, but it is low and does not affect the species distribution of bands. SDS-PAGE profiles show relative
stability of individual bands among these samples. In our previous study (Fyad-Lamèche et al., 2016), on different accessions of these two species, identical results were obtained, confirming our current results. Thus, the seed storage protein SDS-PAGE analysis allows clear identification of each species, even closely related species in the genus Medicago, except in cases where the relationship is not clearly established and the species are easily confused.
The analysis of the total seed storage protein profiles enables identification of batches of seed samples without the need for germination and the determination of which species they belong. They also show whether any seed lot is pure or contaminated with seeds of other species.
In autogamous species, like the two species studied here, a single seed is sufficient to carry out the tests. According to
Table 6. Origin and characteristics of investigated populations.
|
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
*Institut technique de développement des grandes cultures (Sidi-Bel-Abbes. Algeria). Accessions that were studied for their genome size (2C DNA), chromosome number (2n) and seed storage protein are indicated by +. **Accessions for which karyotypes have been performed are indicated by double asterisk.
Cai and Bullen (1992), each seed in a pure line gives rise to the same protein banding pattern. However, in out breeding species, genetic variation may well exist within cultivars to a greater or lesser degree.
Our results clearly show that the two species have clearly distinct profiles, even though they belong to the same section of Leptospirae.
Materials and methods
Plant materials
The origins of the accessions of two studied species (M. polymorpha and M. laciniata) are presented in Table 6. The accessions were provided by ICARDA (International Center for Arid and Dry Area, Aleppo, Syria) and ITGC (Institut Technique des Grandes Cultures. Sidi-Bel-Abbes. Algeria).
Determination of chromosome number and karyotype analysis
The seeds of both species were germinated on wet filter paper in Petri dishes at 4°C for 48h and then at room temperature (22°C) in dark for 3 days. The roots of about 0.5 to 1 cm of length were excised between 8h and 11h in the morning, pre-treated with 8-hydroxyquinoline (0.003M) for 3h at 16°C, and fixed in freshly prepared solution (3:1) of the 95% ethanol: glacial acetic acid (v/v) for at least 24 h. The fixed root meristems were stored at 4°C in the first fixative for few days and then in the 70% ethanol for several months at -20° C. For chromosome count and morphometric analysis of chromosomes, the meristems were hydrolyzed in 1N HCl at 60°C for 12 min, stained in Schiff reagent or acetic-orcein (1% solution in 45% acetic acid) and then squashed in a drop of 1% aceto-carmine (Feulgen and Rosenbach, 1924).
The karyotype was determined by examining at least five well spread metaphase plates obtained from several
accessions of each species (Table 1). The determination of the centromere position and chromosome type was made according to Levan et al. (1964). The asymmetry of the karyotype, based on chromosome morphology, was evaluated by asymmetry index (AsI%) according to the formula of Arano and Saito (1980) AsI%=100 x (sum of the lengths of long arms)/(sum of total chromosome lengths). We also estimate inter-chromosomal and intra-chromosomal asymmetry according to Peruzzi and Eroğlu (2013) and Watanabe et al. (1999). The software used for chromosome measurement was Optika SView.
Genome size evaluation by flow cytometry
The total nuclear DNA amount was assessed using the method of flow cytometry according to Marie and Brown (1993) and Bourgeet al. (2018). The leaf tissue of the target plant was chopped with a razor blade together with leaf tissue of the appropriate internal standard [Petunia hybrid (Hook) Vilm. cv. ‘PxPc6’, 2C = 2.85 pg for M. laciniata and Solanum lycopersicum L. cv. ‘Montfavet 63-5’, 2C = 1.99 pg for M. polymorpha] in 1000 μl of cold Gif Nuclear Isolation Buffer—GNB: 45 mM MgCl2,30 mM sodium citrate, 60 mM MOPS (4-morpholine propane sulphonate, pH = 7), and 1% (w/v) polyvinyl pyrrolidone 10,000, pH 7.2), containing 0.1% (w/v) Triton X–100, supplemented with 5 mM sodium metabisulphite and RNase (2.5 U/mL)(Bourgeet al., 2018). After filtration of the nuclear suspension through a nylon mesh (pore size 50 μm), to remove non-useful tissue fragments, the sample was kept on ice, stained with propidium iodide (stock 1 mg/ml, Sigma-Aldrich, France) to a final concentration of 50 μg/ml and analysed in a flow cytometer (CyFlow SL3, Partec) with a 532 nm 30 mW laser. 2C-value was calculated by multiplying the known DNA content of the internal standard by the peak (modal) position in the histogram of the Medicago species fluorescence intensities and by dividing the result by the peak position of the internal standard. The mean 2C-value from at least five individuals per each studied accession as well as the standard deviation of the mean and its coefficient of variation (%) were calculated.
Electrophoresis of seed storage proteins
The individual extracts were made from the dry seeds in the buffer Tris-HCl0,0625 M of pH=6.8, containing 10% of the glycerol, the sodium dodecyl sulfate (SDS), 2% (w/v) and 2.5% (v/v) of the β-mercaptoethanol. Denaturation of extracts was performed in a water bath for 3 min at 100°C and then centrifuged at 14000 rpm/min for 15 min (Van Geyt and Smed, 1984; Andrews, 1986). The "Protein Biolabs” was used as a molecular weight marker which is a mixture of 12 highly purified reference proteins, easily identifiable as clear bands of 10-230 kDa. The protein electrophoresis under denaturing conditions was performed on the polyacrylamide gel with SDS according to the method described by Laemmli (1970). A discontinuous gel was used, with the stacking gel and the separation gel having a polyacrylamide concentration of 4.5% and 7.5%, respectively. The migration takes place under an intensity of 25 mA the first hour and 30 mA thereafter. The gel was stained in a solution of the methanol and the acetic acid (50%-10%) where the Coomassie blue R250 was dissolved. After gel distaining in the methanol acetic acid solution, and the fixation in the 7% acetic acid solution, a comparison of electrophoretic bands was performed based on the position and thickness, number and their mobility. To avoid ambiguity, the experiment was repeated five times.
Statistical analysis
Student T-tests were used to compare the chromosomes morphometric parameters as well as the DNA content between the two species.
Conclusion
There is no close relationship between the two studied species, and M. laciniata cannot be considered a taxon from which M. polymorpha could have directly evolved. We also point out that the profile of seed storage proteins, which is very stable characteristic, could be used in breeding programmes for varietal identification. Overall, our study provided some valuable information for species identification.
Acknowledgements
We thank SC Brown and M. Bourge for his assistance on the Imagerie-Gif Cytometry core facility of the Gif campus (https://www.i2bc.paris-saclay.fr/bioimaging/cytometry).
References
Abdelguerfi A, Chapot JY, Conesa A (1988) Contribution à l'étude de la répartition des luzernes annuelles spontanées en Algérie selon certains facteurs du milieu. Fourrages (Versailles): 89-106.
Agarwal K, Gupta PK (1983) Cytological studies in the genus Medicago L. Cytologia 48: 781-793.
Andrews AT (1986) Electrophoresis: Theory, techniques and biochemical and clinical applications. Oxford University Press.
Arano H, Saito H (1980) Cytological studies in family Umbelliferae. V. Karyotypes of seven species in subtribe Seselinae. La Kromosomo. (II-17): 471-480.
Badri M, Cheikh NB, Mahjoub A, Abdelly C (2016) Morpho-phenological diversity among natural populations of Medicago polymorpha of different Tunisian ecological areas. Afr J Biotechnol. 15: 1330-1338.
Bauchan G, Elgin JJ (1984) A new chromosome number for the genus Medicago 1. Crop Sci. 24: 193-195.
Bauchan GR, Hossain MA (1999) Constitutive hétérochromatine DNA polymorphisms in diploid medicago sativa ssp. falcate. Genome. 42: 930-935.
Bena G, Lejeune B, Prosperi JM, Olivieri I (1998a) Molecular phylogenetic approach for studying life-history evolution: the ambiguous example of the genus Medicago L. Proceedings of the Royal Society of London. Series B: Biol Sci. 265: 1141-1151.
Bena G, Prosperi JM, LejeuneB, Olivieri I (1998b) Evolution of Annual Species of the Genus Medicago: A Molecular Phylogenetic Approach. Mol Phylogenet Evol. 9: 552-559.
Benmiloud-Mahieddine R, Abirached-Darmency M, Brown SC, Kaid-Harche M, Siljak-Yakovlev S (2011) Genome size and cytogenetic characterization of three Algerian Retama species. Tree Genet Genomes. 7: 987-998.
Bourge M, Brown SC, Siljak-Yakovlev S (2018) Flow cytometry as tool in plant sciences, with emphasis on genome size and ploidy level assessment. Genetics and Applications. 2: 1-12.
Cai Q, Bullen M (1992) Identification of timothy cultivars by SDS-PAGE analysis of seed storage proteins. Can J Plant Sci. 72: 1215-1222.
Ceccarelli M, Minelli S, Maggini F, Cionini PG (1995) Genome size variation in Vicia faba. Heredity. 74(2): 180-187.
Cerbah M, Kevei Z, Siljak-Yakovlev S, Kondorosi E, Kondorosi A, Trinh TH (1999) FISH chromosome mapping allowing karyotype analysis in Medicago truncatula lines Jemalong J5 and R-108-1. Mol Plant Microbe Interact. 12: 947-950.
Chen J, Wu G, Shrestha N, Wu S, Guo W, Yin M, Li A, Liu J, Ren G (2021) Phylogeny and species delimitation of Chinese Medicago (Leguminosae) and its relatives based on molecular and morphological evidence. Front Plant Sci. 11: 619799.
Djafri-Bouallag L, Ourari M, Sahnoune M (2019).) A cytogenetic and pollen study of annual Medicago species from Soummam Valley (Northeastern of Algeria). Acta Bot Croat. 78 (1): 82–90.
Doležel J, Bartoš J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry Part. A 51: 127-128.
Falistocco E (2018) Chromosome investigations in annual Medicago species (Fabaceae) with emphasis on the origin of the polyploid Medicago rugosa and Medicago scutellata. Plant Biosyst. 153: 235-241.
Falistocco E, Falcinelli M, Veronesi F (1995) Karyotype and C-banding pattern of mitotic chromosomes in alfalfa, Medicago sativa L. Plant Breed. 114: 451-453.
Feulgen R, Rosenbach H (1924) Chemische Nachweise von Nukleinsäuren vom Typ der Thymonukleinsäure und die darauf beruhende selektive Färbung von Zellkernen in mikroskopischen Präparaten. F Hoppe- Seyler’sZeitschr. PhysiolChem. 135: 203-248.
Fyad-Lamèche Z (1998) Variabilité des protéines de réserve des graines de populations d'espèces annuelles de Medicago. Acta Bot Gall. 145: 199-217.
Fyad-Lamèche F-Z, Iantcheva A, Siljak-Yakovlev S, Brown SC (2016) Chromosome number, genome size, seed storage protein profile and competence for direct somatic embryo formation in Algerian annual Medicago species. Plant Cell Tissue Organ Cult (PCTOC). 124: 531-540.
Garnatje T, Vilatersana R, Susanna A, Valles J, Siljak-Yakovlev S (2004) Contribution to the karyological knowledge of Echinops (Asteraceae, Cardueae) and related genera. Bot J Linn Soc. 145: 337-344.
Gillies C (1971) Alfalfa Chromosomes. III. Medicago Glomerata Balb. Pachytene Karyotype 1. Crop Sci. 11: 463-464.
Greilhuber J, Doležel J, Lysák MA, Bennett MD (2005) The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann Bot. 95: 255-260.
Heyn CC (1963)The annual species of Medicago. Editeur: Jerusalem [ISR]: Hebrew University, 154 pp.
Hidalgo O, Garcia-Jacas N, Garnatje T, Susanna A, Siljak-Yakovlev S (2007) Karyological evolution in Rhaponticum Vaill. (Asteraceae, Cardueae) and related genera. Bot J Linn Soc. 153: 193-201.
Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences. 97: 6603-6607.
Krochko JE, Bewley JD (1988) Use of electrophoretic techniques in determining the composition of seed storage proteins in alfalfa. Electrophoresis. 9: 751-763.
Krochko JE, Bewley JD (2000) Seed storage proteins in cultivars and subspecies of alfalfa (Medicago sativa L.). Seed Sci Res. 10: 423-434.
Laamari R, Ochatt S, Ferchichi A (2016) Ploidy level, genome size and genetic variability among a collection of Medicago sativa L. Gabsi as revealed by flow cytometry. J New Sci. 33.
Ladizinsky G, Hymowitz T (1979) Seed protein electrophoresis in taxonomic and evolutionary studies. Theor Appl Genet. 54: 145-151.
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680-685.
Lesins K, Gillies C (1972) Taxonomy and cytogenetics of Medicago. Alfalfa. Sci Technol. 15: 53-86.
Lesins K, Lesins I, Gillies C (1970) Medicago murex with 2n= 16 and 2n= 14 chromosome complements. Chromosoma. 30: 109-122.
Lesins KA, Lesins I (1979) Genus Medicago (Leguminosae): A taxonomic study. Dr. W. Junk Publishers, The Hague, Netherlands.
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas. 52: 201-220.
Mariani A, Falistocco E (1990) Chromosome studies in 2n = 14 and 2n = 16 types of Medicago murex. Genome. 33: 159-163.
Mariani A, Pupilli F, Calderini O (1996) Cytological and molecular analysis of annual species of the genus Medicago. Canad J Bot. 74: 299-307.
Marie D, Brown SC (1993) A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biol Cell 78: 41-51.
Panigrahi J, Kumar D, Mishra M, Mishra R, Jena P (2007) Genomic relationships among 11 species in the genus Cajanus as revealed by seed protein (albumin and globulin) polymorphisms. Plant Biotechnol Rep. 1: 109-116.
Peruzzi L, Eroğlu HE (2013) Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenetics. 7(1):1-9.
Prosperi J, Guy P, Genier G, Angevain M (1995) Les luzernes ou le genre Medicago. INRA Editions.
Prosperi J, Jenczewski E, Ronfort J (2000) The mielgas: wild Spanish populations of alfalfa. Results of ten years of researches. Lucerne and medics for the XXI Century. Proceedings XIII Eucarpia Medicago spp. Group Meeting, Perugia, Italy, 13-16 September 1999. Universita di Perugia, pp 1-10.
Quiros CF, Bauchan GR (1988) The genus Medicago and the origin of the Medicago sativa comp. Alfalfa and Alfalfa Improvement. 29: 93-124.
Rajpal VR, Singh A, Kumar A, Sharma S, Raina SN (2022) Deciphering species relationships between the wild and cultivated species of genus Medicago based on seed protein profiles. Proceedings of the National Academy of Sciences, India Section B: Biol Sci. 92: 533-540.
Sadeghian S, Hejazi SH (2014) Cytogenetic studies in some species of Medicago L. in Iran. Eur J Biolsci. 73: 21-30.
Siljak-Yakovlev S (1996) La dysploïdie et l'évolution du caryotype. Bocconea. 5: 210-220.
Singh A, Raina SN, Rajpal VR, Singh AK (2018) Seed protein fraction electrophoresis in peanut (Arachis hypogaea L.) accessions and wild species. Physiol Mol Biol Plants. 24: 465-481.
Small E (2011) Alfalfa and relatives: evolution and classification of Medicago. NRC research press.
Small E, Bauchan GR (1984) Chromosome numbers of the Medicago sativa complex in Turkey. Canad J Bot. 62: 749-752.
Small E, Jomphe M (1989) A synopsis of the genus Medicago (Leguminosae). Canad J Bot. 67: 3260-3294.
Steele KP, Ickert‐Bond SM, Zarre S, Wojciechowski MF (2010) Phylogeny and character evolution in Medicago (Leguminosae): Evidence from analyses of plastid trnK/matK and nuclear GA3ox1 sequences. Am J Bot. 97: 1142-1155.
Van Geyt J, Smed E (1984) Polymorphism of some marker enzymes of the sugarbeet (Beta vulgaris L.) investigated by polyacrylamide gel electrophoresis and starch gel electrophoresis. Z. Pflanzenzticht. 92: 295-308.
Watanabe K, Yahara T, Denda T, Kosuge K (1999) Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae): statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. J Plant Res. 112: 145-161.
Yahia N, Fyad-Lameche FZ (2003) Évaluation de la variabilité de jeunes plants de Medicago soumis à un régime de basse température. Acta Bot Gallica. 150: 3-17.
Yin L, Shang-Zhi H, Jia-Rui F (1998) One and two-dimensional polyacrylamide gel electrophoretic analysis of seed polypeptide composition of peanut cultivars. Acta Bot Sin. 40.