

ISSN:1835-2707
Aust J Crop Sci. 18(08):500-507 (2024)
https://doi.org/10.21475/ajcs.24.18.08.pne196
Alternative plant protection using an indigenous bacterium (Stutzerimonas nitrititolerans) isolated from agricultural soil of potato fields in Western Algeria to biodegrade common pesticides
Nahla Bekenniche1*, Arezki Ait Abdeslam2,3, Djamila Maghnia3,4
1Laboratory of Food Technology and nutrition, Department of Biology, Faculty of Nature and Life Sciences, University Abdelhamid Ibn Badis Mostaganem, 27000, Algeria
2Department of Biology, Faculty of Nature and Life Sciences, University of Mustapha Stambouli Mascara -29000, Algeria
3Laboratory of Experimental Toxicology, BioDePollution and PhytoRemediation (BTE-BD-PR), Department of Biology, SNV Faculty, Oran 1 Ahmed Ben Bella University -31000, Oran, Algeria
4Department of Biology, Faculty of Nature and Life Sciences, University Abdelhamid Ibn Badis Mostaganem, 27000, Algeria
Abstract
Nowadays, it is highly important to find ways to degrade pesticides residues from the environment. We studied degradation of carbamate, copper sulphate - Cymoxanil and Cycloxydim by using an indeginous bacterium Stutzerimonas nitrititolerans. This bacteria uses pesticides as source of carbon and solubilize them by their enzymatic machinery. Experiments were conducted using mineral salt medium (MSM) on agar supplemented with two different pesticides as sole carbon and energy source: carbamate and acetamiprid. These organic pesticides contain carbon (example of carbamate: NH2COOH). Growth of the bacteria could suggest that it has utilized the pesticide as its sole energy source, indicating its ability to degrade the pesticide. The effects of different factors on pesticides degradation were investigated such us pesticide tolerance. The bacteria were tested for the ability to tolerate high concentrations of pesticides to optimize the degradation conditions. The treatments consisted of carbon-free mineral salts medium (MSM) supplemented with pesticides that are usually used during cultivation of potato such as acetamiprid, carbamate, copper sulphate - Cymoxanil and Cycloxydim at increasing concentrations (70, 100, 200 and 500mgl-1). Bacterial growth was monitored by daily measurement of the optical density at 600 nm on UV-1600 Spectrophotometer. Only one strain (Labeled PF2 of Stutzerimonas nitrititolerans) showed potential to tolerate and mitigate the tested pesticides at high concentrations. Further, a phenotypic characterization was carried, and 16S rRNA gene sequencing analysis performed to identify the PF2 strain. Pesticides did not show any significant inhibitory effect on the isolate PF2. Comparison of sequences with GenBank database gave similarity with Stutzerimonas nitrititolerans with 99.74% of homology. A phylogenetic tree of Stutzerimonas nitrititolerans was constructed to show relationship to closely related bacteria. The potential and efficiency of the strain PF2 using pesticides can be considered as a qualitative leap in the field of pesticide bioremediation in agricultural soils.
Keywords: Agricultural soil, Potato crops, pesticides,bioremediation,Stutzerimonas nitrititolerans, 16S rRNA gene.
Abbreviation: MSM_ mineral salt medium; PF2_ Stutzerimonas nitrititolerans stain’s code; OD_ optical density; pH_ potential of hydrogen.
Introduction
The potato (Solanum tuberosum L.) has become a key model for food consumption. The excessive use of pesticides by human on this crop has increase potato yields but caused soil pollution. Pesticide residues are toxic and effective against pests such as insects, nematodes, and rats, as well as functioning as insecticides, herbicides, and fungicides (Tamm et al., 2022). Various studies have shown that microbial activity is the most significant factor in xenobiotics degradation, although physical-chemical factors like moisture content, temperature, pH, pesticide formulation, and organic carbon content can also influence the process (Asamba et al., 2022). The principle is to use the toxic component as the only source of energy and at the same time reduce the toxiciy of compond (Ortiz-Hernández et al., 2013).
Bioremediation is advantageous due to capability of microbes for detoxification of the environmental pollutants, e.g. Pseudomonas sp., Bacillus sp, Klebsiella sp, Pandoraea sp, Phanerochaete Chrysosporium, Mycobacterium sp. The pesticides are eventually transformed or degraded by the microorganism as a carbon source, nitrogen source, any other mineral source or a final electron acceptor in respiratory chain.
For example, Achromobacter xylosoxidans strain CS5 was able to utilize both endosulfan and endosulfan sulfate as sulfur as well as carbon source and energy source resulting in complete mineralization of endosulfan via hydrolytic pathway (Wen et al., 2009).
Carbamates are non-persistent pesticides in the environment but the World Health Organization has classified them as toxic and their use is restricted due to their crucial degradation and their toxicity for living beings (Mustapha et al., 2019). Acetamiprid’s utilization presents a risk of ecological contamination. The excessive application of acetamiprid to improve crop yield has resulted in the detection of residual traces in various natural sources (Xu et al., 2020). Cymoxanil is a fungicide, type of acetamide molecule that exhibits local systemic effects. The use of microorganisms in the field of biodegradation is not new. The principle is to use the toxic component as the only source of energy and at the same time reduce the toxiciy of compound (Ortiz-Hernández et al., 2013). Soil bacteria continuously exposed to toxic chemicals such as pesticides can develop an ability to degrade these chemicals. The search for new soil bacteria with the potential to degrade pesticides remains the best solution to combat the toxicity of soils and crops. This approach is less expensive and respects the environment (Huang et al., 2021). Microorganisms have specific enzymes that have an essential role in pesticide bioremediation (Mali et al., 2022). Microbes that were frequently observed in the process of pesticide bioremediation include Pseudomonas sp., Bacillus sp, Flavobacterium sp., Mycobacterium sp., Sphingomonas sp. (Ruomeng et al., 2023). In bioremediation, living microorganism is used to remove pesticides by breaking them down into less toxic forms. This depends on the type of pesticide, the environmental matrix, and the organisms present in the ecosystem. This study focuses on indigenous bacteria that were isolated from agricultural soil of potato exposed for more than 20 years to high concentrations of pesticides. The strain has been identified as Stutzerimonas nitrititolerans. With the aim of improving their ability to degrade these chemicals, a primary investigation was conducted with monitoring of bacterial survival under different growth conditions. Biodegradation was studied in detail and this isolate was found to grow in a selective medium supplemented with different pesticides. This study aimed at deciphering the biological control to mitigate the use of these compounds in the agricultural soil of potato and obtain organic crops, by isolating bacteria with potential biodegradation for pollutants trapped in the soil.
Results
Soil characteristics
The soil sample was a loamy soil, ideal for growing potatoes because they provide good drainage while retaining enough moisture and nutrients for plant growth. The soil’s pH was 5.5 which is an appropriate level which ensures that essential nutrients are readily accessible to the potato plants. The weather conditions were as follow: a temperature of 6°C and at moderate wind speeds of 6 km/h, humidity level of 70% and a significant rainfall.
Morphological and biochemical characterization
The isolated Stutzerimonas nitrititolerans bateria (Labeled PF2) was circular, yellow, smooth and shiny colony with regular edges. Microscopic observation showed a rod-shaped form and gram negative (Table1).
PF2 was motile, non-spore forming, facultatively anaerobic, catalase and oxidase positive. It could grow at 10, 20, 30, 40°C with pH 4, 6, 7, 10 and NaCl percentages of 4%, 5%, 6%, 7%.
Carbohydrate consumption
Positive for glucose, fructose, mannose and negative for xylose, galactose, lactose, trehalose, ribose, arabinose. PF2 was negative for arginine dihydrolase, indol and H2S production. Antibiotic resistance was observed for gentamicin (6mm diameter), ampicillin (8mm diameter), Erythromycin (11mm diameter), Vancomycin (6mm diameter) and Streptomycin (10mm diameter) (Table 1).
Pesticide tolerance test on MSM agar
The evaluation of strain PF2 tolerance to carbamate and acetamiprid at higher pesticide concentrations showed considerable bacterial colonies at 70, 100, 200 mgl-1 after 4 days of incubation. While at a concentration of 500 mgl-1, we had to wait until the 6th day of incubation to have few colonies. The strain PF2 exposed to chemicals was considered tolerant to these pesticides.
Growth of PF2 at high pesticide’s concentrations
All measurements were performed in triplicate and results are reported on microbial growth curves of MSM cultures supplemented with different concentrations of pesticides. Chemicals were chosen in accordance with the farmer's treatment protocol on the potato crops. The MSM medium supplemented with pesticides but without inoculum was considered as negative control (Fig 1). A significant increasing growth (p<0.001) of the strain PF2 was noted with all pesticides at different concentrations used in the experience.
Molecular characterization of PF2
Isolate PF2 was identified based on the similarity to sequences in the GenBank. The analysis of the sequences obtained by the National Center for Biotechnology Information (NCBI) database. BLAST results identified PF2 isolate as Stutzerimonas nitrititolerans with 99.74% of homology (Fig 2). Phylogenetic tree showing the relationship of Stutzerimonas nitrititolerans to closely related bacteria was constructed using MEGA11 (MEGA Software Company). Strains were clustered together based on the similarity of sequences. The sequences were aligned using the Neighbor-joining method. Bootstrap values were calculated and indicated (1000 replications). The tree's scale and the unit for branch lengths as evolutionary distances were 0.05.
Nucleotide sequence accession number
Data associated with this study has been deposited at NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession number: (GenBank accession nos. OR690900).
Discussion
The aerobic metabolism of the studied soil with the four pesticides tested separately and at increasing concentrations of 50, 100 and 200 mg L-1 did not have an inhibitory effect on the PF2 isolate. The growth of this bacteria in MSM-cycloxidime at 500 mg L was low, compared to its growth in MSM-Cooper sulfate-cymoxanil. By observing the increasing growth curves, we could conclude that bacteria are better adapted with Cooper sulfate-cymoxanil. The Copper sulfate has microbial activity against a wide range of microorganisms commonly used to prevent the crop phytopathogenic (Fortunato et al., 2021). Therefore, the tolerance of our isolate to this phytosanitary compound in the soil is positive for our research if we wish to involve it in the field of bioremediation and biocultures. Carbamate alone and acetamiprid alone at 500 mg L-1 in MSM was degraded efficiently and quickly by PF2 as sole carbon source. This is reflected by the period of adaptability that PF2 had previously on the tolerance medium MSM added by aarbamate and acetamiprid together. Fast-growing species may have an advantage in their use in organic crops compared to their competitors (Khan et al., 2023). It is crucial to gradually increase the dose of pesticides to allow bacteria better adaptability with the pollutant. The effect of temperature, pH and salt content are physiological parameters that can influence cell growth in agriculture. PF2 was able to grow in an acidic to neutral environment without difficulty knowing that the cultivar pH of potato is ranged from about 5.5 to 6.2 (Kiszonasand Bamberg, 2009). For the temperature, we were able to observe rapid growth of the strain at increasing temperatures. Dismaying the homology of the sequences which is 99.74% % for the species Stutzerimonas nitrititolerans and the phylogenetic tree, Stutzerimonas is a bacterial genus that belongs to the family Pseudomonadaceae. It is often associated with denitrification processes, meaning it can convert nitrate and nitrite into nitrogen gas under anaerobic conditions. Stutzerimonas species, including Stutzerimonas nitrititolerans, are known for their ability to tolerate and use nitrite as a nitrogen source in their metabolic processes. A closer homology of this species was observed with genus Pseudomonas sp. This is why a reclassification has been proposed. In the phylogenetic part, the name Pseudomonas nitrititolerans sp. nov. type strain: GL14T (=CGMCC 1.13874T=NBRC 113853T) was proposed. It was a novel strain isolated from the nitrification-denitrification bioreactor (Peng at al., 2019). The genus Stutzerimonas was created recently and 10 species of Pseudomonas were transferred to it, whereas new name and new combinations in the genus Stutzerimonas gen. nov. were born (Lalucat et al., 2022). A description of the new combinations in the genus Stutzerimonas gen. nov. was given (Gomila et al., 2022). The strain Pseudomonas nitrititolerans sp. was proposed and assigned to Stutzerimonas as Stutzerimonas nitrititolerans. The genus Pseudomonas sp. was selected for tolerance to pesticides and its antioxidative enzymes against the herbicides (Rovida et al., 2021). This potency may be due to different enzymes (hydrolase, esterase) and genes like ophB. The strain Stutzerimonas nitrititolerans PF2- OR690900 can be considered as a new species in the field of biodegradation of pesticides, isolated from potato crops. The combined results of the above phylogenetic, and phenotypic analyses suggested that strain PF2 represents the first species of the genus Stutzerimonas isolated from potato crops and the first strain tolerating pesticides at increasing concentrations. By introducing these bacteria into contaminated environments, it is possible to facilitate the breakdown of pesticides into less harmful substances, thus mitigating environmental damage.
Materials and methods
Study site
The study was caried out in Mesra, Mostaganem, Algeria (Latitude: 35.8373, Longitude: 0.169785, 35° 50′ 14″ North, 0° 10′ 11″ East). The localisation of collection site of potato soils is shown in Figure 3. The climate of Mesra is dry and hot semi-arid (https://weatherspark.com/y/42391/Average-Weather-in-Mostaganem-Algeria-Year-Round#Sections-BestTime) (Figure 3).
Sample collection
Agricultural soil of a potato field, treated with different pesticides for long time, were collected. The collection was done in January at 6°C. The sample was obtained at a depth of about 15cm of the soil and transferred into a bag aseptically and stored at 4°C.
History of treatments
An investigation was done and a conversation with the farmer informed us of the history of using pesticides. The method of treatment carried out on this potato agricultural soil has been the same for 10 years. Briefly, first, the soil was treated with the granulated mineral nitrogen fertilizer ammonitrate 33.5%. NPK fertilizer was used to enrich the soil. Second treatment was biological by adding organic cow manure. During cultivation, several kinds of pesticides have also been used throughout the years to ensure maximum yield. In this study, the choice of pesticides was based on the history of treatments that the soil had undergone. All pesticides used in experiments below were purchased from Agrichem Algeria (acetamiprid, carbamate, copper sulphate – Cymoxanil, Cycloxydim). The commercialized pesticides (Insecticide, Bactericide – Fungicide, Acaricides, and Herbicide) which have been used for our experiences were Rustilan, Decafate, Lannate, and Focus Ultra, respectively (Table 2).
Culture media and strain isolation
For targeted isolation of pesticide-tolerant bacteria, a mineral salt medium (MSM) supplemented with two pesticides as sole carbon source, acetamiprid and carbamate were used. Soil sample (10g) was cultivated in 90ml mineral salt medium (MSM) containing (in gl-1): 4.3 K2HPO4 (Merck); 3.4 KH2PO4 (Merck); 2.0 (NH4)2SO4 (Merck); 0.16 MgCl2 (Merck); 0.001 MnCl24H2O (Merck); 0.0006 FeSO47H2O (Sigma); 0.026 CaCl22H2O (Sigma); and 0.02 Na2MoO42H2O (Merck). Pesticides (Acetamiprid and Carbamate) filtered with millipore filters (0.45 µm) were added at a concentration of 50mgl-1 each. Culture was incubated at 30°C with shaking at 200 rpm for 15 days. One milliliter of this culture was diluted then inoculated on MSM agar (1.5% agar) sprayed with the same pesticides carbamate and acetamiprid. The Petri dishes were incubated for 48h to 72h at 30°C (Kimura et al., 2018). Bacterial colony was observed after 24 hours of incubation on MSM agar. Bacterial strain was inoculated directly in Luria–Bertani (LB) broth and incubated for 24h. The overnight culture of the strain (PF2) was centrifuged and the pellet resuspended in conservation medium (0.5% yeast extract, 1% tryptone, 1% NaCl, pH 7.0 supplemented with 20% glycerol).
Tolerance test on solid medium
For higher pesticide concentrations, concentrations of 70, 100, 200 and 500 mgl-1 of carbamate and acetamiprid were added to MSM agar. The strain was inoculated in each concentration and the agar plates incubated for 7 days at 30°C. MSM agar medium without bacterial inoculation was used as a control (Xu et al., 2020).
Pesticides kinetics
The strain PF2 was tested in a liquid MSM medium supplemented with a single pesticide as a carbon source. Pesticides were selected following the history of agricultural soil treatment: Acetamiprid, Carbamate, Copper sulphate - Cymoxanil and Cycloxydim at increasing concentrations.
The PF2 preculture preparation
PF2 was precultured in LB for 48 h at 30°C with shaking at 220 rpm, then the suspension centrifuged at 8000 rpm for 10 min. The supernatant was discarded and the bacterial pellet was washed with sterile MSM three times. This pellet was resuspended in MSM and stored at 4 ◦C for further study. The preculture of PF2 was tested with increasing concentrations of each pesticide (50, 70, 100, 200 and 500mgl-1) Bacterial growth was monitored for 12 days under the same incubation conditions by measuring the optical density at 600 nm on UV-1600 Spectrophotometer. MSM with pesticides and without bacterial inoculation was used as the control (Wang et al., 2013). Each treatment was set in triplicate.
Statistical analysis
All statistical tests were performed using the commercial software Statistical Package for Social Sciences (SPSS version 23.0). The bacterial growth on different pesticide’s concentrations was performed using one-way parametric analysis of variance (ANOVA). Significant differences (P value ≤ 0.05) were determined by multiple comparison test, Tukey.
Phenotypic characterization
Cell morphology, gram and spore forming were observed by microscopy. Growth at different temperature, pH, and NaCl concentration were determined after incubation on a nutrient broth at 30 °C.
Biochemical characteristics
Biochemical tests were performed on a 24 hours old culture grown on nutrient medium plate at 30˚C. Catalase, oxidase, H2S, indol, nitrate reduction, respiratory mode and cetrimide were performed. Other tests such as enzymes for sugars, ADH and antibiotic resistance were carried out to get as close as possible to the species.
Antimicrobial susceptibility
The method used is diffusion in MH medium using the conditions recommended by CA-SFM / EUCAST (2022). The antibiotics used were: Gentamicin (10IU), Ampicillin (10IU), Erythromycin (15µg), Vancomycin (30µg) and Streptomycin (10µg) (Cypress Diagnostics, Belgium) (CA-SFM/ EUCAST, 2022).
Molecular analysis
Extraction of DNA
The commercial kits Wizard®, ReliaPrep™ DNA Purification Kit from Promega was used to extract genomic DNA from bacteria PF2. Following the manufacturer's recommendations, the steps consisted of preparing a 24h bacterial suspension of PH2 in nutrient broth. 2ml of the suspension were centrifuged at 10,000 rpm for 2 minutes. 1ml of bacterial suspension was kept at -20ºC to repeat the experiment if necessary and 1 mL was centrifuged for 2 minutes at 13,000 rpm. The supernatant was removed and the pellet collected and resuspended in 480 μL of TE buffer (10 mM Tris HCl pH 7.6;1 mM EDTA), then with a micropipette 600 μL of nucleic lysis solution and 10 μL of Proteinase K (PK) were added and resuspended meticulously (De Brito et al., 2022).
PCR amplification
Primers used in bacterial DNA amplification of the 16S rRNA gene were 27F (5'AGTTTGATCCTGGCTCAG-3') and 1492R (5'GTTACCTTGTTACGACTTC-3'). The program respected in the Polymerase Chain Reaction (PCR) is as follows: first hold of temperature at 95°C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30s, annealing at 55 °C for 45s and extension at 72 °C for 90s. A final elongation was done at 72°C for 10 min (Bekenniche et al., 2021).
Phylogenetic analysis
The BLAST was used to find the identical sequence in the database of nucleotide sequences from NCBI (http://www.ncbi.nlm.nih.gov/BLAST). The most abundant unique sequence for the isolate PF2 was searched against the NCBI 16S microbial database using BLASTn, with the argument- max_target_seqs 20. The results obtained were sorted first by their value, then by their score and the taxonomy of the highest score sequence was reported. To show the phylogenetic positions of strain PF2 and other related species, homologous sequences were added in Molecular Evolutionary Genetics Analysis (MEGA 11.0)(Kumar et al., 2018), available at https://www.megasoftware.net/. Distances from each branch were calculated using: “The Neighbor-Joining Method” by Jukes-Cantor (Saitou and Nei, 1987). The representative sequence of the strain PF2 was selected and submitted to the NCBI website (www.submit.ncbi.nlm.nih.gov/subs) to have the GenBank accession number.
Conclusion
New biological approaches have provided promising information on the degradation of pesticides. Several bacterial genera have been discovered from agricultural soils polluted by pesticides, which have developed enzymatic systems involved in the metabolism of pesticides. Strain PF2- Stutzerimonas nitrititolerans has demonstrated a high tolerance to different pesticides and various stressors. This strain has a high adaptability to environments exposed to pesticides, which is an asset for the future of organic cropping. However, it is important to note that the effectiveness of Stutzerimonas nitrititolerans in pesticide biodegradation depends on various factors, including the specific pesticides present, the concentration of contaminants, environmental conditions, and the overall microbial community composition. Research in this area is ongoing, and further studies would be needed to assess the practical applicability and efficiency of Stutzerimonas nitrititolerans in biodegrading pesticides under different conditions.
Conflict of interests
The authors declare no conflict of interest.
References
Bekenniche N, Ait Abdeslam A, Nemmiche S, Ait Saada D, Cheriguenen A, Bekada A M A(2021) Molecular identification of crude oil-degrading bacteria isolated from polluted sites in Algeria. South Asian J Exp Biol. 11 (2): 164-171.https://doi.org/10.38150/sajeb.11(2).p164-171
CA-SFM/ EUCAST (2022) European Committee on Antimicrobial Susceptibility Testing. Societé francaise de microbiologie. https://www.sfm-microbiologie.org/wp-content/uploads/2022/05/CASFM2022_V1.0.pdf.
De Brito OA, dos Santos FA, Al Yafawi TT Saraiva CRN, da Silva ROM, Leandro LMG, de Aquino PEA, de Sousa Júnior DL, Leandro MKNS (2022) Comparing protocols of DNA extraction from Escherichia coli: Analysis of purity and concentration by gel electrophoresis.Baghdad Journal of Biochemistry and Applied Biological Sciences. 3(02) :133-144.https://doi.org/10.47419/bjbabs.v3i02.123
Fortunato G, Vaz-Moreira I, Nunes OC, Manaia CM (2021) Effect of copper and zinc as sulfate or nitrate salts on soil microbiome dynamics and blaVIM-positive Pseudomonas aeruginosa survival. Journal of Hazardous Materials. 415 : 125631.https://doi.org/10.1016/j.jhazmat.2021.125631
Gomila M, Mulet M, García-Valdés E, Lalucat J (2022) Genome-based taxonomy of the genus Stutzerimonas and proposal of S. frequens sp. nov. and S. degradans sp. nov. and emended descriptions of S. perfectomarina and S. chloritidismutans microorganisms. 10:1363. https://doi.org/10.3390/microorganisms10071363
Huang Y, Zhang W, Pang S (2021) Insights into the microbial degradation and catalytic mechanisms of chlorpyrifos. Environ Res. 194:110660. https://doi.org/10.1016/j.envres.2020.110660
Khan B, Jamil I, Mirani ZA, Hassan ZUl, Azhar A, Galani S (2023) Investigation and application of a novel Pseudomonas aeruginosa (KIBGE-AB9) for the biodegradation of malathion in agriculture soil. International Journal Of Agriculture and Biology. 30:65‒7. DOI: 10.17957/IJAB/15.2059
Kimura W T, Suenaga H, Fujihara H, Futagami T, Goto M, Hanada S, Hirose J (2018) Pseudomonas furukawaii sp. nov., a polychlorinated biphenyldegrading bacterium isolated from biphenyl-contaminated soil in Japan. Int J Syst Evol Microbiol. 68(5):1429-1435. https://doi.org/10.1099/ijsem.0.002670
Kiszonas AM, and Bamberg J (2009) Survey of tuber pH variation in potato (Solanum) species. American Journal of Potato Research. 87(2):167-176. https://doi.org/10.1007/s12230-009-9120-0
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35:1547-1549.https://doi.org/10.1093/molbev/msy096
Lalucat J, Gomila M, Mulet M, Zaruma A, García-Valdés E (2022) Past, present and future of the boundaries of the Pseudomonas genus: Proposal of Stutzerimonas gen. Nov. Systematic and Applied Microbiology. 45:126289. https://doi.org/10.1016/j.syapm.2021.126289
Mali H, Shah C, Darshan H, Patel Trivedi U, Subramanian RB (2022) Degradation insight of organophosphate pesticide chlorpyrifos through novel intermediate 2,6‑dihydroxypyridine by Arthrobacter sp. HM01. Bioresources and Bioprocessing. 9:31. https://doi.org/10.1186/s40643-022-00515-5
Mustapha MU, Halimoon N, Johar WW, Abd Shukor MY (2019) An overview on biodegradation of carbamate pesticides by soil bacteria. Pertanika Journal of Science Technology. 27:2. https://www.researchgate.net/publication/332601003
Ortiz-Hernández M L, Sánchez-Salinas E, Castrejón Godínez M L, Dantan González E, Popoca Ursino UC (2013) Mechanisms and strategies for pesticide biodegradation: Opportunity for waste, soils and water cleaning. Revista internacional De Contamination Ambiental. 2985-104. http://www.redalyc.org/articulo.oa?id=37028958005
Peng JS, Liu Y, Yan L, Hou TT, Liu HC, Zhou YG, Liu ZP (2019) Pseudomonas nitrititolerans sp. nov., a nitrite-tolerant denitrifying bacterium isolated from a nitrification/denitrification bioreactor. Int J Syst Evol Microbiol. 69:2471–2476. https://doi.org/10.1099/ijsem.0.003516
Rovida AFdS, Costa G, Santos MI, Silva CR, Freitas PNN, Oliveira EP, Pileggi SAV, Olchanheski RL and Pileggi M (2021) Herbicides tolerance in a Pseudomonas strain is associated with metabolic plasticity of antioxidative enzymes regardless of selection. Front Microbiol. 12:673211. Front. Microbiol. 673211.https://doi.org/10.3389/fmicb.2021.673211
Ruomeng B, Meihao O, Siru Z, Shichen G, Yixian Z, Junhong C, Ruijie M, Yuan L, Gezhi X, Xingyu C, Shiyi Z, Aihui Z, Baishan F (2023) Degradation strategies of pesticide residue: From chemicals to synthetic biology. Synthetic and Systems Biotechnology 8: 302–313. https://doi.org/10.1016/j.synbio.2023.03.005
Saitou N and Nei M (1987) The neighbor-joining method: A new meth-od for reconstructing phylogenetic trees. Molecular Biology and Evolution. 4:406-25. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Tamm L, Thuerig B, Apostolov S, Blogg H, Borgo E, Corneo PE, Fittje S, et al (2022) Use of copper-based fungicides in organic agriculture in twelve european countries. Agronomy. 12: 673. https://doi.org/10.3390/agronomy12030673
Wang G, Chen X, Yue W, Zhang H, Li F, Xiong M (2013) Microbial degradation of acetamiprid by ochrobactrum sp. D-12 isolated from contaminated soil. PLoS ONE. 8(12): e82603. https://doi.org/10.1371/journal.pone.0082603
Xu B, Xue R, Zhou J, Wen X, Shi Z, Chen M, Xin F, Zhang W, Dong W, Jiang M (2020) Characterization of acetamiprid biodegradation by the microbial consortium ACE-3 enriched from contaminated soil. Front Microbiol. 11:1429. https://doi.org/10.3389/fmicb.2020.01429
| Table 1. Differential characteristic of strain PF2 | |
|---|---|
| Morphological Characteristics | |
| colony shape | Circular- Smooth- Shiny |
| Colour | Yellow |
| Edges | Regular |
Microscopic observation
|
|
| Cell form | Bacilli |
| Gram Stain | - |
| Motility | + |
| Spore | - |
| Respiratory tests | |
| Oxydase | + |
| Catalase | + |
| Nitrate reduction | + |
| Respiratory type | facultatively anaerobic |
| ADH | - |
| H2S production | - |
| Fluorecent Pigment | - |
| Growth on different environmental conditions | |
| Growth Temperature (°C) | |
| 10 | + |
| 20 | + |
| 30 | + |
| 40 | + |
| Growth NaCl (%) | |
| 4 | + |
| 5 | + |
| 6 | + |
| 7 | + |
| 8 | - |
| Growth pH | |
| 4 | + |
| 6 | + |
| 7 | + |
| 10 | - |
| Antibiotic resistance (inhibition zone : mm) | |
| Gentamycin | 6 |
| Ampicillin | 8 |
| Erythromycin | 11 |
| Vancomycin | 6 |
| Streptomycin | 10 |
| Assimilation of | |
| Glucose | + |
| Fructose | + |
| Mannose | + |
| Xylose | - |
| Galactose | - |
| Lactose | - |
| Trehalose | - |
| Ribose | - |
| Arabinose | - |
| +, tested positive/utilized as substrate; 2, tested negative/not utilized as substrate. | |
| Table 2. Different kinds of pesticides used in potato crops | |||
|---|---|---|---|
| Kinds of Pesticides | Commercial Name | ||
| Insecticide | Acetamipride | Rustilan |  |
| Bactericide – Fongicide | copper sulphate - Cymoxanil | Decafate | |
| Acaricides | Carbamate | Lannate | |
| Herbicide | Cycloxydim | Focus Ultra | |
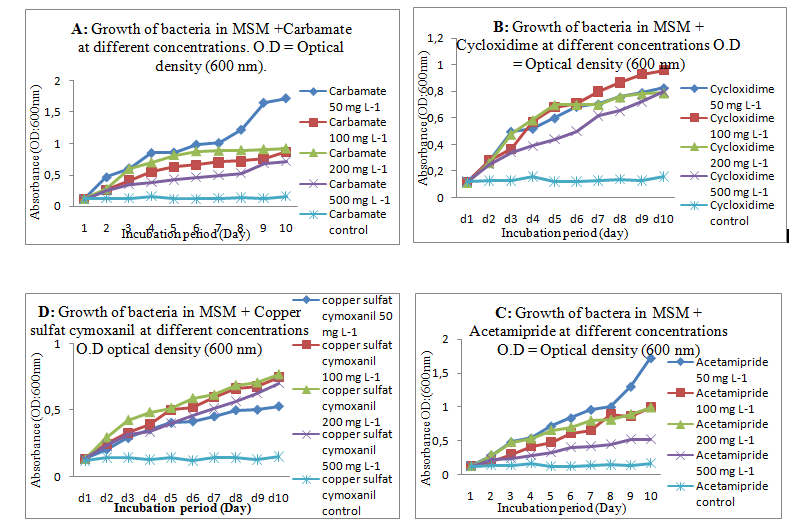
Figure 1. Growth of strain PF2 in MSM media at increasing concentrations of pesticides. O.D (600 nm.A: MSM +Carbamate.B: MSM + Cycloxidim.C: MSM + Acetamiprid. D:MSM + Copper sulfate cymoxanil).Control : MSM medium + pesticides.
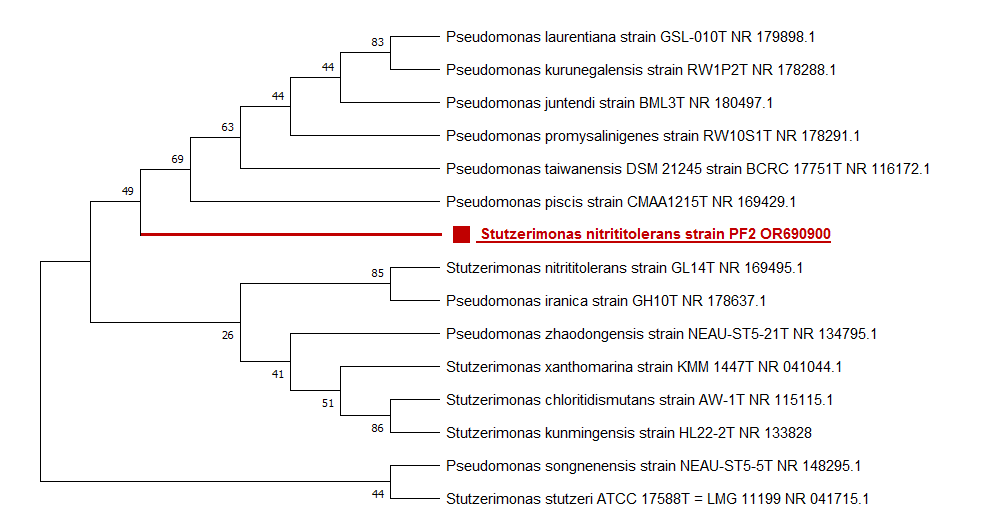
Figure 2. Neighbor-joining tree based on concatenated sequences of the 16S rRNA of strain PF2 and other related Stutzerimonas species. Bootstrap values >50% (based on 1000 replications) are shown at branch points. Filled square indicate branches of strainPF2 Stutzerimonas nitrititolerans. Bar 0.05 substitutions per nucleotide site.
Figure 3. Localisation of collection site (a, b, c: Google maps 2024), and Agricultural soil of potato (d).